Enhanced proofreading governs CRISPR-Cas9 targeting accuracy
- PMID: 28931002
- PMCID: PMC5918688
- DOI: 10.1038/nature24268
Enhanced proofreading governs CRISPR-Cas9 targeting accuracy
Abstract
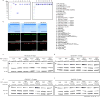
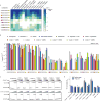
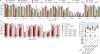
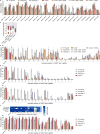
The RNA-guided CRISPR-Cas9 nuclease from Streptococcus pyogenes (SpCas9) has been widely repurposed for genome editing. High-fidelity (SpCas9-HF1) and enhanced specificity (eSpCas9(1.1)) variants exhibit substantially reduced off-target cleavage in human cells, but the mechanism of target discrimination and the potential to further improve fidelity are unknown. Here, using single-molecule Förster resonance energy transfer experiments, we show that both SpCas9-HF1 and eSpCas9(1.1) are trapped in an inactive state when bound to mismatched targets. We find that a non-catalytic domain within Cas9, REC3, recognizes target complementarity and governs the HNH nuclease to regulate overall catalytic competence. Exploiting this observation, we design a new hyper-accurate Cas9 variant (HypaCas9) that demonstrates high genome-wide specificity without compromising on-target activity in human cells. These results offer a more comprehensive model to rationalize and modify the balance between target recognition and nuclease activation for precision genome editing.
Conflict of interest statement
J.K.J. has financial interests in Beacon Genomics, Beam Therapeutics, Editas Medicine, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, and Intellia Therapeutics; a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics and Driver; and executive director of the Innovative Genomics Institute at UC Berkeley and UCSF. S.H.S. is an employee of Caribou Biosciences, Inc. S.H.S., J.S.C. and J.A.D. are inventors on a patent application entitled “Reporter Cas9 variants and methods of use thereof” (PCT/US2016/036754), filed by The Regents of the University of California. B.P.K. and J.K.J. are inventors on a patent application entitled “Engineered CRISPR-Cas9 nucleases” (US 15/060,424), filed by The General Hospital Corporation. J.S.C, Y.S.D., B.P.K, A.Y., J.K.J. and J.A.D. have filed a patent application related to this work through The General Hospital Corporation and The Regents of the University of California.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

