SAXS and stability studies of iron-induced oligomers of bacterial frataxin CyaY
- PMID: 28931050
- PMCID: PMC5607177
- DOI: 10.1371/journal.pone.0184961
SAXS and stability studies of iron-induced oligomers of bacterial frataxin CyaY
Abstract
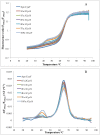
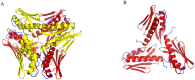
Frataxin is a highly conserved protein found in both prokaryotes and eukaryotes. It is involved in several central functions in cells, which include iron delivery to biochemical processes, such as heme synthesis, assembly of iron-sulfur clusters (ISC), storage of surplus iron in conditions of iron overload, and repair of ISC in aconitase. Frataxin from different organisms has been shown to undergo iron-dependent oligomerization. At least two different classes of oligomers, with different modes of oligomer packing and stabilization, have been identified. Here, we continue our efforts to explore the factors that control the oligomerization of frataxin from different organisms, and focus on E. coli frataxin CyaY. Using small-angle X-ray scattering (SAXS), we show that higher iron-to-protein ratios lead to larger oligomeric species, and that oligomerization proceeds in a linear fashion as a results of iron oxidation. Native mass spectrometry and online size-exclusion chromatography combined with SAXS show that a dimer is the most common form of CyaY in the presence of iron at atmospheric conditions. Modeling of the dimer using the SAXS data confirms the earlier proposed head-to-tail packing arrangement of monomers. This packing mode brings several conserved acidic residues into close proximity to each other, creating an environment for metal ion binding and possibly even mineralization. Together with negative-stain electron microscopy, the experiments also show that trimers, tetramers, pentamers, and presumably higher-order oligomers may exist in solution. Nano-differential scanning fluorimetry shows that the oligomers have limited stability and may easily dissociate at elevated temperatures. The factors affecting the possible oligomerization mode are discussed.
Conflict of interest statement
Figures








Similar articles
-
Iron-induced oligomerization of human FXN81-210 and bacterial CyaY frataxin and the effect of iron chelators.PLoS One. 2017 Dec 4;12(12):e0188937. doi: 10.1371/journal.pone.0188937. eCollection 2017. PLoS One. 2017. PMID: 29200434 Free PMC article.
-
The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.J Biol Chem. 2013 Mar 22;288(12):8156-8167. doi: 10.1074/jbc.M112.442285. Epub 2013 Jan 23. J Biol Chem. 2013. PMID: 23344952 Free PMC article.
-
Oligomerization propensity and flexibility of yeast frataxin studied by X-ray crystallography and small-angle X-ray scattering.J Mol Biol. 2011 Dec 16;414(5):783-97. doi: 10.1016/j.jmb.2011.10.034. Epub 2011 Oct 25. J Mol Biol. 2011. PMID: 22051511 Free PMC article.
-
Frataxin Structure and Function.Subcell Biochem. 2019;93:393-438. doi: 10.1007/978-3-030-28151-9_13. Subcell Biochem. 2019. PMID: 31939159 Review.
-
Assembly of Fe/S proteins in bacterial systems: Biochemistry of the bacterial ISC system.Biochim Biophys Acta. 2015 Jun;1853(6):1436-47. doi: 10.1016/j.bbamcr.2014.12.009. Epub 2014 Dec 13. Biochim Biophys Acta. 2015. PMID: 25510311 Review.
References
-
- Babcock M, deSilva D, Oaks R, Davis Kaplan S, Jiralerspong S, Montermini L, et al. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276(5319):1709–12. . - PubMed
-
- Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM, Chasteen ND. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J Mol Biol. 2004;341(2):605–15. doi: 10.1016/j.jmb.2004.05.072 - DOI - PubMed
-
- He YN, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A, et al. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43(51):16254–62. doi: 10.1021/bi0488193 - DOI - PMC - PubMed
-
- Cavadini P, O'Neill HA, Benada O, Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Hum Mol Gen. 2002;11(3):217–27. . - PubMed
-
- Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millan-Pacheco C, Pastor N, et al. The structure and function of frataxin. Crit Rev Bioch Mol Biol. 2006;41(5):269–91. doi: 10.1080/10409230600846058 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

