Identification and molecular characterization of a metagenome-derived L-lysine decarboxylase gene from subtropical soil microorganisms
- PMID: 28931053
- PMCID: PMC5607190
- DOI: 10.1371/journal.pone.0185060
Identification and molecular characterization of a metagenome-derived L-lysine decarboxylase gene from subtropical soil microorganisms
Abstract
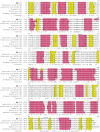
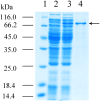
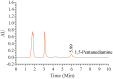
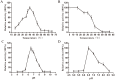
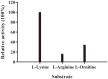
L-lysine decarboxylase (LDC, EC 4.1.1.18) is a key enzyme in the decarboxylation of L-lysine to 1,5-pentanediamine and efficiently contributes significance to biosynthetic capability. Metagenomic technology is a shortcut approach used to obtain new genes from uncultured microorganisms. In this study, a subtropical soil metagenomic library was constructed, and a putative LDC gene named ldc1E was isolated by function-based screening strategy through the indication of pH change by L-lysine decarboxylation. Amino acid sequence comparison and homology modeling indicated the close relation between Ldc1E and other putative LDCs. Multiple sequence alignment analysis revealed that Ldc1E contained a highly conserved motif Ser-X-His-Lys (Pxl), and molecular docking results showed that this motif was located in the active site and could combine with the cofactor pyridoxal 5'-phosphate. The ldc1E gene was subcloned into the pET-30a(+) vector and highly expressed in Escherichia coli BL21 (DE3) pLysS. The recombinant protein was purified to homogeneity. The maximum activity of Ldc1E occurred at pH 6.5 and 40°C using L-lysine monohydrochloride as the substrate. Recombinant Ldc1E had apparent Km, kcat, and kcat/Km values of 1.08±0.16 mM, 5.09±0.63 s-1, and 4.73×103 s-1 M-1, respectively. The specific activity of Ldc1E was 1.53±0.06 U mg-1 protein. Identifying a metagenome-derived LDC gene provided a rational reference for further gene modifications in industrial applications.
Conflict of interest statement
Figures


References
-
- Bunsupa S, Katayama K, Ikeura E, Oikawa A, Toyooka K, Saito K, et al. Lysine decarboxylase catalyzes the first step of quinolizidine alkaloid biosynthesis and coevolved with alkaloid production in leguminosae. Plant Cell. 2012;24(3): 1202–1216. doi: 10.1105/tpc.112.095885 - DOI - PMC - PubMed
-
- Bhatia SK, Bhatia RK, Yang YH. Biosynthesis of polyesters and polyamide building blocks using microbial fermentation and biotransformation. Rev Environ Sci Biotechnol. 2016;15(4): 639–663. doi: 10.1007/s11157-016-9415-9 - DOI
-
- Kind S, Kreye S, Wittmann C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab Eng. 2011;13(5): 617–627. doi: 10.1016/j.ymben.2011.07.006 - DOI - PubMed
-
- Kind S, Neubauer S, Becker J, Yamamoto M, Völkert M, Abendroth Gv, et al. From zero to hero-production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab Eng. 2014;25: 113–123. doi: 10.1016/j.ymben.2014.05.007 - DOI - PubMed
-
- Tsuge Y, Kawaguchi H, Sasaki K, Kondo A. Engineering cell factories for producing building block chemicals for bio-polymer synthesis. Microb Cell Fact. 2016;15: 19 doi: 10.1186/s12934-016-0411-0 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

