Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma
- PMID: 28931080
- PMCID: PMC5607158
- DOI: 10.1371/journal.pone.0184969
Low serum level of miR-485-3p predicts poor survival in patients with glioblastoma
Abstract
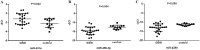
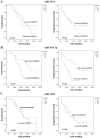
MicroRNAs (miRNAs) are short noncoding RNAs that play critical roles in human malignancies and can be used as biomarkers for cancer. Until now, a number of biomarkers for prognosis of glioblastoma (GBM) have been reported in tumor tissues but only a few biomarkers in circulating fluid. Using a custom microarray, we previously identified 19 differentially expressed miRNAs in serum of patients with GBM. In this study, we investigated whether 3 of the 19 miRNAs in serum could be used as prognostic biomarkers for patients with GBM. We first validated the serum levels of 3 candidate miRNAs in an independent cohort of 24 GBM patients and 12 healthy volunteers by real-time quantitative reverse transcription PCR (qRT-PCR), and then evaluated the prognostic value of these miRNAs in a total of 36 GBM patients. The results show that the serum levels of the 3 miRNAs (miR-451a, miR-485-3p and miR-4298) determined by qRT-PCR are significantly different between 24 GBM patients and 12 healthy volunteers (all P <0.05) and are in concordance with the results of microarray analysis. High serum level of miR-451a is correlated with positive tumor O(6)-methylguanine-DNA methyltransferase (MGMT) expression (P = 0.040). Survival analysis showed that low serum miR-485-3p level is associated with poor progression-free survival (PFS) (P < 0.004) and overall survival (OS) (P < 0.023). Furthermore, univariate and multivariate Cox analyses demonstrated that that serum miR-485-3p expression is a significant independent prognostic factor for PFS and OS in GBM patients. In conclusion, serum miR-485-3p level is reduced and might be a potential prognostic biomarker in GBM patients.
Conflict of interest statement
Figures
Similar articles
-
Up-regulation of miR-370-3p restores glioblastoma multiforme sensitivity to temozolomide by influencing MGMT expression.Sci Rep. 2016 Sep 6;6:32972. doi: 10.1038/srep32972. Sci Rep. 2016. PMID: 27595933 Free PMC article.
-
A 4-miRNAs signature predicts survival in glioblastoma multiforme patients.Cancer Biomark. 2017 Dec 6;20(4):443-452. doi: 10.3233/CBM-170205. Cancer Biomark. 2017. PMID: 28869437
-
Reduced circulating microRNA-203 predicts poor prognosis for glioblastoma.Cancer Biomark. 2017 Dec 6;20(4):521-526. doi: 10.3233/CBM-170335. Cancer Biomark. 2017. PMID: 28946553
-
The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients' survival: a meta-analysis.World J Surg Oncol. 2016 Oct 12;14(1):261. doi: 10.1186/s12957-016-1012-4. World J Surg Oncol. 2016. PMID: 27733166 Free PMC article. Review.
-
The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials.J Cell Physiol. 2018 Jan;233(1):378-386. doi: 10.1002/jcp.25896. Epub 2017 May 16. J Cell Physiol. 2018. PMID: 28266716 Review.
Cited by
-
The plasma miRNome and venous thromboembolism in high-grade glioma: miRNA Sequencing of a nested case-control cohort.J Cell Mol Med. 2024 Apr;28(8):e18149. doi: 10.1111/jcmm.18149. J Cell Mol Med. 2024. PMID: 38613361 Free PMC article.
-
Involvement of Differentially Expressed microRNAs in the PEGylated Liposome Encapsulated 188Rhenium-Mediated Suppression of Orthotopic Hypopharyngeal Tumor.Molecules. 2020 Aug 8;25(16):3609. doi: 10.3390/molecules25163609. Molecules. 2020. PMID: 32784458 Free PMC article.
-
[Long-chain non-coding RNA MALAT1 regulates paclitaxel resistance of breast cancer cells by targeting miR-485-3p].Nan Fang Yi Ke Da Xue Xue Bao. 2020 May 30;40(5):698-702. doi: 10.12122/j.issn.1673-4254.2020.05.13. Nan Fang Yi Ke Da Xue Xue Bao. 2020. PMID: 32897218 Free PMC article. Chinese.
-
Long Noncoding RNA SNHG7, a Molecular Sponge for microRNA-485, Promotes the Aggressive Behavior of Cervical Cancer by Regulating PAK4.Onco Targets Ther. 2020 Jan 23;13:685-699. doi: 10.2147/OTT.S232542. eCollection 2020. Onco Targets Ther. 2020. Retraction in: Onco Targets Ther. 2021 Sep 09;14:4741-4742. doi: 10.2147/OTT.S337553. PMID: 32158221 Free PMC article. Retracted.
-
Glioblastoma Multiforme miRNA based Comprehensive Study to Validate Phytochemicals for Effective Treatment against Deadly Tumour through In Silico Evaluation.Microrna. 2024;13(3):240-250. doi: 10.2174/0122115366302365240618122812. Microrna. 2024. PMID: 38982916
References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352(10):987–96. Epub 2005/03/11. doi: 10.1056/NEJMoa043330 . - DOI - PubMed
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. doi: 10.1016/S1470-2045(09)70025-7 . - DOI - PubMed
-
- Karsy M, Neil JA, Guan J, Mahan MA, Colman H, Jensen RL. A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg Focus. 2015;38(3):E4 doi: 10.3171/2015.1.FOCUS14755 . - DOI - PubMed
-
- De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–35. doi: 10.1093/annonc/mdu239 . - DOI - PMC - PubMed
-
- Garcia-Romero N, Carrion-Navarro J, Esteban-Rubio S, Lazaro-Ibanez E, Peris-Celda M, Alonso MM, et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget. 2017;8(1):1416–28. doi: 10.18632/oncotarget.13635 . - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

