Isolation and Molecular Characterization of the Transformer Gene From Bactrocera cucurbitae (Diptera: Tephritidae)
- PMID: 28931159
- PMCID: PMC5469387
- DOI: 10.1093/jisesa/iex031
Isolation and Molecular Characterization of the Transformer Gene From Bactrocera cucurbitae (Diptera: Tephritidae)
Abstract
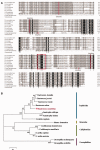
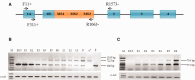
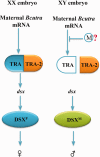
transformer (tra) is a switch gene of sex determination in many insects, particularly in Dipterans. However, the sex determination pathway in Bactrocera cucurbitae (Coquillett), a very destructive pest on earth, remains largely uncharacterized. In this study, we have isolated and characterized one female-specific and two male-specific transcripts of the tra gene (Bcutra) of B. cucurbitae. The genomic structure of Bcutra has been determined and the presence of multiple conserved Transformer (TRA)/TRA-2 binding sites in Bcutra has been found. BcuTRA is highly conservative with its homologues in other tephritid fruit flies. Gene expression analysis of Bcutra at different developmental stages demonstrates that the female transcript of Bcutra appears earlier than the male counterparts, indicating that the maternal TRA is inherited in eggs and might play a role in the regulation of TRA expression. The conservation of protein sequence and sex-specific splicing of Bcutra and its expression patterns during development suggest that Bcutra is probably the master gene of sex determination of B. cucurbitae. Isolation of Bcutra will facilitate the development of a genetic sexing strain for its biological control.
Keywords: Bactrocera cucurbitae (Coquillett); Bcutra; sex determination; sex-specific splicing; transformer.
© The Authors 2017. Published by Oxford University Press on behalf of Entomological Society of America.
Figures

Similar articles
-
Molecular characterizations of two cytochrome P450 genes encoding CYP6A41 and CYP6EK1 from the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae).Arch Insect Biochem Physiol. 2012 Jan;79(1):31-46. doi: 10.1002/arch.21003. Arch Insect Biochem Physiol. 2012. PMID: 23589219
-
The alternative splicing of BdTai and its involvement in the development of Bactrocera dorsalis (Hendel).J Insect Physiol. 2017 Aug;101:132-141. doi: 10.1016/j.jinsphys.2017.07.012. Epub 2017 Jul 24. J Insect Physiol. 2017. PMID: 28750998
-
The role of the transformer gene in sex determination and reproduction in the tephritid fruit fly, Bactrocera dorsalis (Hendel).Genetica. 2015 Dec;143(6):717-27. doi: 10.1007/s10709-015-9869-7. Genetica. 2015. PMID: 26481008
-
Developmental Biology of Zeugodacus cucurbitae (Diptera: Tephritidae) in Three Cucurbitaceous Hosts at Different Temperature Regimes.J Insect Sci. 2015 Nov 20;15(1):160. doi: 10.1093/jisesa/iev141. Print 2015. J Insect Sci. 2015. PMID: 26589874 Free PMC article. Review.
-
Early embryonic transcriptomes of Zeugodacus tau provide insight into sex determination and differentiation genes.Insect Sci. 2022 Jun;29(3):915-931. doi: 10.1111/1744-7917.12974. Epub 2021 Oct 29. Insect Sci. 2022. PMID: 34553826 Review.
Cited by
-
A Novel Solid Artificial Diet for Zeugodacus cucurbitae (Diptera: Tephritidae) Larvae With Fitness Parameters Assessed by Two-Sex Life Table.J Insect Sci. 2020 Jul 1;20(4):21. doi: 10.1093/jisesa/ieaa058. J Insect Sci. 2020. PMID: 32809019 Free PMC article.
-
Identification of sex determination genes and their evolution in Phlebotominae sand flies (Diptera, Nematocera).BMC Genomics. 2019 Jun 25;20(1):522. doi: 10.1186/s12864-019-5898-4. BMC Genomics. 2019. PMID: 31238870 Free PMC article.
References
-
- Belote J. M., McKeown M., Boggs R. T., Ohkawa R., Sosnowski B. A.. 1989. Molecular genetics of transformer, a genetic switch controlling sexual differentiation in Drosophila. Dev. Genet. 10: 143–154. - PubMed
-
- Boggs R. T., Gregor P., Idriss S., Belote J. M., McKeown M.. 1987. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 50: 739–747. - PubMed
-
- Bopp D., Saccone G., Beye M.. 2014. Sex determination in insects: variations on a common theme. Sexual Dev. 8: 20–28. - PubMed
-
- Burtis K. C., Baker B. S.. 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 56: 997–1010. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

