Fasciclin-calcareous corpuscle binary complex mediated protein-protein interactions in Taenia solium metacestode
- PMID: 28931431
- PMCID: PMC5606126
- DOI: 10.1186/s13071-017-2359-2
Fasciclin-calcareous corpuscle binary complex mediated protein-protein interactions in Taenia solium metacestode
Abstract
Background: Neurocysticercosis (NC) caused by Taenia solium metacestode (TsM) is a serious neurological disease of global concern. Diverse bioactive molecules involved in the long-term survival of TsM might contribute to disease progression. Fasciclin (Fas) is an extracellular protein that mediates adhesion, migration and differentiation of cells by interacting with other molecules. We hypothesized that TsMFas might bind to calcareous corpuscle (CC) through its adhesive property and participate in crucial protein-protein interactions, thus contributing to the creation of a symbiotic interactome network.
Methods: Two paralogous TsMFas (TsMFas1 and TsMFas2) were isolated, and their molecular properties were characterized. The co-localization pattern of TsMFas1 and TsMFas2 with CC was determined. CC-TsMFas binary complex was generated by incubating CC with recombinant proteins (rTsMFas1 and 2). In vitro binding assay of CC-rTsMFas1 or CC-rTsMFas2 binary complex with TsM cellular proteins extracted from scolex and neck was conducted. Their binding partners were identified through proteomic analysis. Integrated protein-protein interaction networks were established.
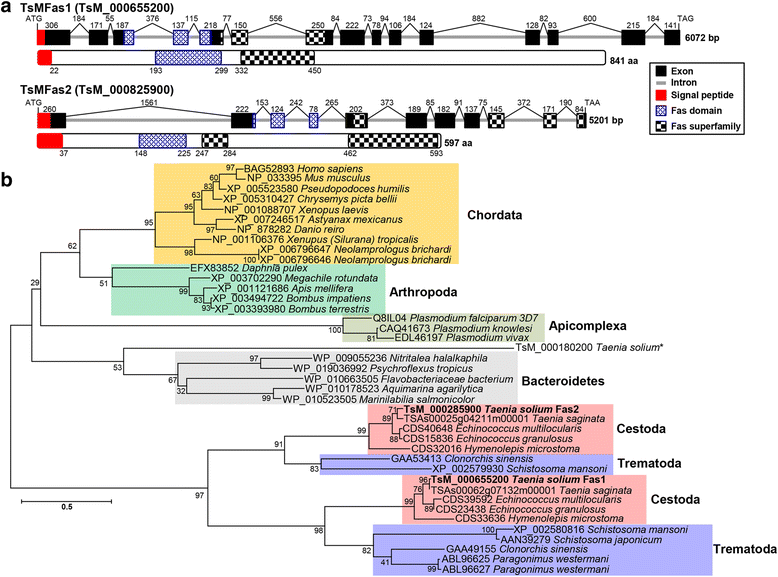
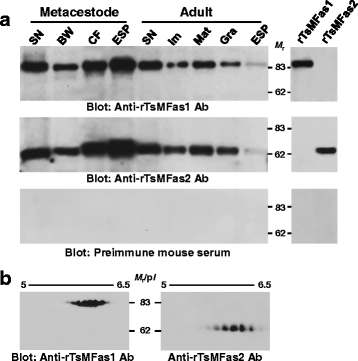
Results: TsMFas1 (6072 bp long) was composed of 15 exons (841 amino acid polypeptide) interrupted by 14 introns. TsMFas2 (5201 bp long) comprised of 11 exons (597 amino acids) and 10 intervening introns. These proteins displayed 22% amino acid sequence identity to each other, but tightly conserved Fas-related domains. Several isoforms of Fas1 and Fas2 proteins might have been expressed through post-translational modifications. They showed adhesion activity with other cells. TsMFas proteins were largely distributed in parenchymal regions of the scolex and bladder wall. These molecules were co-localized with CC, a unique organelle found in platyhelminths. Subsequent proteome analysis of CC-Fas binary complex mediated protein-protein interactions revealed seven protein ligands in the TsM cellular proteins. Their functions were mainly segregated into carbohydrate metabolism (enolase, phosphoenolpyruvate carboxykinase, phosphoglycerate kinase and glyceraldehyde 3-phosphate dehydrogenase) and cytoskeleton/cellular motility (actin, paramyosin and innexin nuc-9). Those proteins had direct (physical) and/or indirect (functional) relationships along with their biochemical properties and biological roles.
Conclusion: Protein repertoires strongly suggest that TsMFas and CC may symbiotically mediate protein-protein interactions during biological processes to maintain efficacious homeostatic functions and ensure the prolonged survival of TsM in the host.
Keywords: Calcareous corpuscle; Carbohydrate metabolizing enzyme; Extracellular matrix; Fasciclin; Neurocysticercosis; Protein-protein interactions; Taenia solium metacestode.
Conflict of interest statement
Ethics approval and consent to participate
Study protocols involving the collection of adult worms from patients and
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

