Hypomethylated domain-enriched DNA motifs prepattern the accessible nucleosome organization in teleosts
- PMID: 28931432
- PMCID: PMC5607494
- DOI: 10.1186/s13072-017-0152-2
Hypomethylated domain-enriched DNA motifs prepattern the accessible nucleosome organization in teleosts
Abstract
Background: Gene promoters in vertebrate genomes show distinct chromatin features such as stably positioned nucleosome array and DNA hypomethylation. The nucleosomes are known to have certain sequence preferences, and the prediction of nucleosome positioning from DNA sequence has been successful in some organisms such as yeast. However, at gene promoters where nucleosomes are much more stably positioned than in other regions, the sequence-based model has failed to work well, and sequence-independent mechanisms have been proposed.
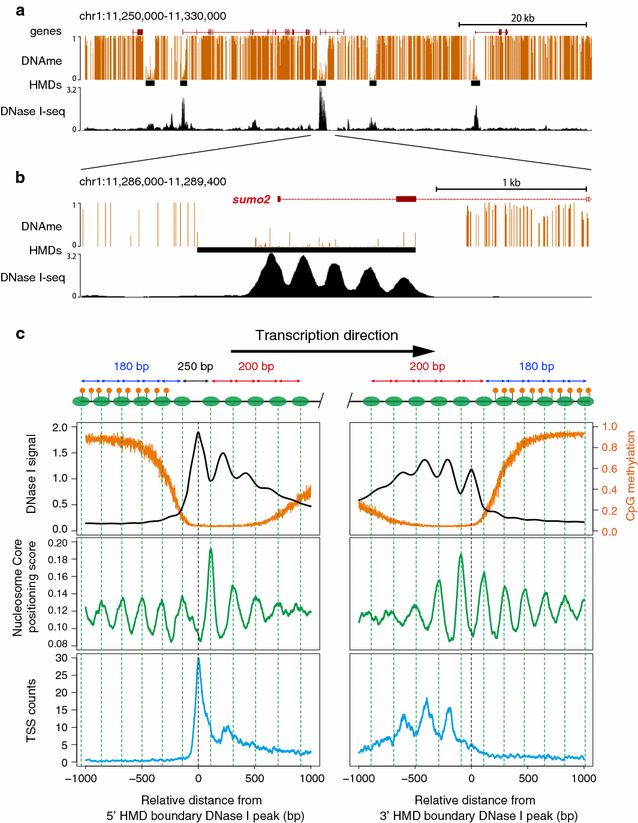
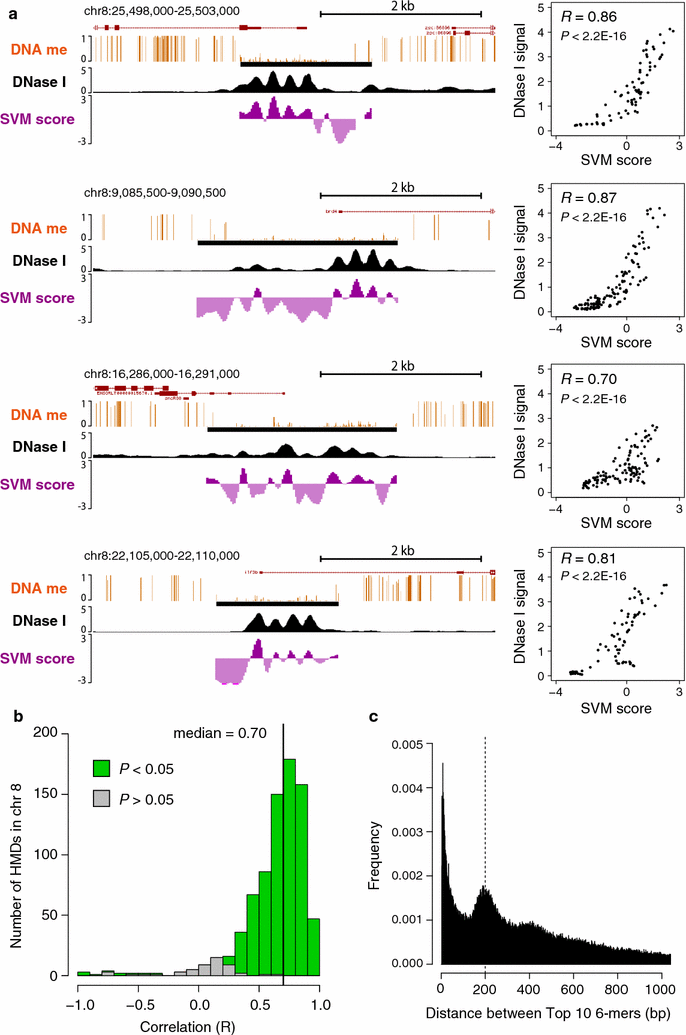
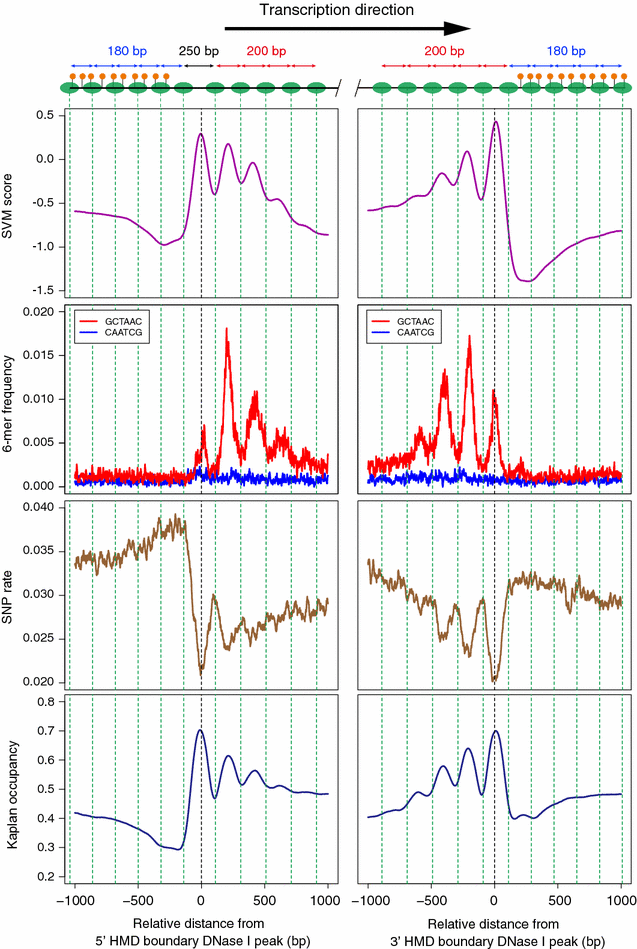
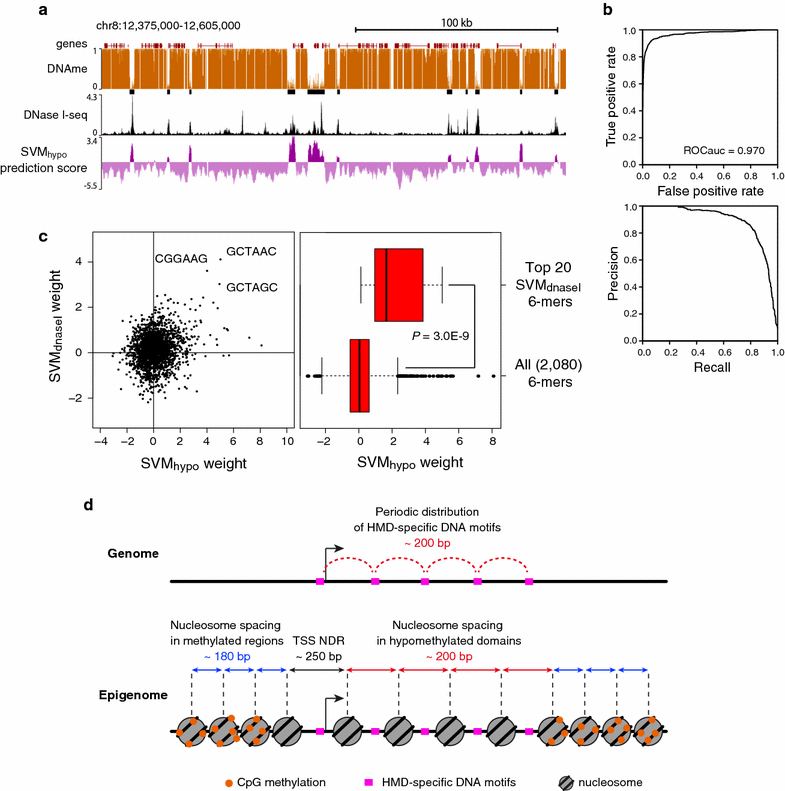
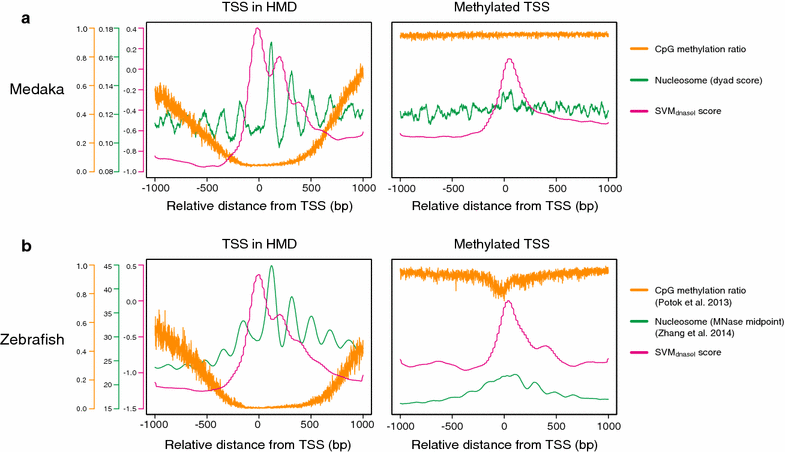
Results: Using DNase I-seq in medaka embryos, we demonstrated that hypomethylated domains (HMDs) specifically possess accessible nucleosome organization with longer linkers, and we reassessed the DNA sequence preference for nucleosome positioning in these specific regions. Remarkably, we found with a supervised machine learning algorithm, k-mer SVM, that nucleosome positioning in HMDs is accurately predictable from DNA sequence alone. Specific short sequences (6-mers) that contribute to the prediction are specifically enriched in HMDs and distribute periodically with approximately 200-bp intervals which prepattern the position of accessible linkers. Surprisingly, the sequence preference of the nucleosome and linker in HMDs is opposite from that reported previously. Furthermore, the periodicity of specific motifs at hypomethylated promoters was conserved in zebrafish.
Conclusion: This study reveals strong link between nucleosome positioning and DNA sequence at vertebrate promoters, and we propose hypomethylated DNA-specific regulation of nucleosome positioning.
Keywords: DNA methylation; DNA sequence; Nucleosome positioning; Vertebrate.
Figures
Similar articles
-
Controls of nucleosome positioning in the human genome.PLoS Genet. 2012;8(11):e1003036. doi: 10.1371/journal.pgen.1003036. Epub 2012 Nov 15. PLoS Genet. 2012. PMID: 23166509 Free PMC article.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
-
Nucleosome Organization in Human Embryonic Stem Cells.PLoS One. 2015 Aug 25;10(8):e0136314. doi: 10.1371/journal.pone.0136314. eCollection 2015. PLoS One. 2015. PMID: 26305225 Free PMC article.
-
Ubiquitous human 'master' origins of replication are encoded in the DNA sequence via a local enrichment in nucleosome excluding energy barriers.J Phys Condens Matter. 2015 Feb 18;27(6):064102. doi: 10.1088/0953-8984/27/6/064102. Epub 2015 Jan 7. J Phys Condens Matter. 2015. PMID: 25563930
-
Coupling between Sequence-Mediated Nucleosome Organization and Genome Evolution.Genes (Basel). 2021 Jun 1;12(6):851. doi: 10.3390/genes12060851. Genes (Basel). 2021. PMID: 34205881 Free PMC article. Review.
Cited by
-
Recapitulation-like developmental transitions of chromatin accessibility in vertebrates.Zoological Lett. 2019 Nov 14;5:33. doi: 10.1186/s40851-019-0148-9. eCollection 2019. Zoological Lett. 2019. PMID: 31807314 Free PMC article.
-
Targeted in vivo epigenome editing of H3K27me3.Epigenetics Chromatin. 2019 Mar 13;12(1):17. doi: 10.1186/s13072-019-0263-z. Epigenetics Chromatin. 2019. PMID: 30871638 Free PMC article.
-
Unlinking the methylome pattern from nucleotide sequence, revealed by large-scale in vivo genome engineering and methylome editing in medaka fish.PLoS Genet. 2017 Dec 21;13(12):e1007123. doi: 10.1371/journal.pgen.1007123. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29267279 Free PMC article.
-
Exploring the Link between Nucleosome Occupancy and DNA Methylation.Front Genet. 2018 Jan 12;8:232. doi: 10.3389/fgene.2017.00232. eCollection 2017. Front Genet. 2018. PMID: 29379519 Free PMC article.
-
Deciphering the genetic code of DNA methylation.Brief Bioinform. 2021 Sep 2;22(5):bbaa424. doi: 10.1093/bib/bbaa424. Brief Bioinform. 2021. PMID: 33432324 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

