Nonrandom γ-TuNA-dependent spatial pattern of microtubule nucleation at the Golgi
- PMID: 28931596
- PMCID: PMC5687021
- DOI: 10.1091/mbc.E17-06-0425
Nonrandom γ-TuNA-dependent spatial pattern of microtubule nucleation at the Golgi
Abstract
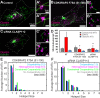
Noncentrosomal microtubule (MT) nucleation at the Golgi generates MT network asymmetry in motile vertebrate cells. Investigating the Golgi-derived MT (GDMT) distribution, we find that MT asymmetry arises from nonrandom nucleation sites at the Golgi (hotspots). Using computational simulations, we propose two plausible mechanistic models of GDMT nucleation leading to this phenotype. In the "cooperativity" model, formation of a single GDMT promotes further nucleation at the same site. In the "heterogeneous Golgi" model, MT nucleation is dramatically up-regulated at discrete and sparse locations within the Golgi. While MT clustering in hotspots is equally well described by both models, simulating MT length distributions within the cooperativity model fits the data better. Investigating the molecular mechanism underlying hotspot formation, we have found that hotspots are significantly smaller than a Golgi subdomain positive for scaffolding protein AKAP450, which is thought to recruit GDMT nucleation factors. We have further probed potential roles of known GDMT-promoting molecules, including γ-TuRC-mediated nucleation activator (γ-TuNA) domain-containing proteins and MT stabilizer CLASPs. While both γ-TuNA inhibition and lack of CLASPs resulted in drastically decreased GDMT nucleation, computational modeling revealed that only γ-TuNA inhibition suppressed hotspot formation. We conclude that hotspots require γ-TuNA activity, which facilitates clustered GDMT nucleation at distinct Golgi sites.
© 2017 Sanders et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
Similar articles
-
Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network.Dev Cell. 2007 Jun;12(6):917-30. doi: 10.1016/j.devcel.2007.04.002. Dev Cell. 2007. PMID: 17543864 Free PMC article.
-
How Microtubules Build the Spindle Branch by Branch.Annu Rev Cell Dev Biol. 2022 Oct 6;38:1-23. doi: 10.1146/annurev-cellbio-120420-114559. Epub 2022 Jun 27. Annu Rev Cell Dev Biol. 2022. PMID: 35759800 Free PMC article. Review.
-
Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130.EMBO J. 2009 Apr 22;28(8):1016-28. doi: 10.1038/emboj.2009.47. Epub 2009 Feb 26. EMBO J. 2009. PMID: 19242490 Free PMC article.
-
Detection and Analysis of Microtubule Nucleator γ-Tubulin Ring Complex.Methods Mol Biol. 2023;2557:543-558. doi: 10.1007/978-1-0716-2639-9_32. Methods Mol Biol. 2023. PMID: 36512236
-
Microtubule nucleation for spindle assembly: one molecule at a time.Trends Biochem Sci. 2023 Sep;48(9):761-775. doi: 10.1016/j.tibs.2023.06.004. Epub 2023 Jul 21. Trends Biochem Sci. 2023. PMID: 37482516 Free PMC article. Review.
Cited by
-
Microtubule nucleation and γTuRC centrosome localization in interphase cells require ch-TOG.Nat Commun. 2023 Jan 26;14(1):289. doi: 10.1038/s41467-023-35955-w. Nat Commun. 2023. PMID: 36702836 Free PMC article.
-
Molecular insight into how γ-TuRC makes microtubules.J Cell Sci. 2021 Jul 15;134(14):jcs245464. doi: 10.1242/jcs.245464. Epub 2021 Jul 23. J Cell Sci. 2021. PMID: 34297125 Free PMC article. Review.
-
Divergent Contribution of the Golgi Apparatus to Microtubule Organization in Related Cell Lines.Int J Mol Sci. 2022 Dec 19;23(24):16178. doi: 10.3390/ijms232416178. Int J Mol Sci. 2022. PMID: 36555819 Free PMC article.
-
A New Look at the Functional Organization of the Golgi Ribbon.Front Cell Dev Biol. 2019 Aug 21;7:171. doi: 10.3389/fcell.2019.00171. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31497600 Free PMC article.
-
Regulation of Glucose-Dependent Golgi-Derived Microtubules by cAMP/EPAC2 Promotes Secretory Vesicle Biogenesis in Pancreatic β Cells.Curr Biol. 2019 Jul 22;29(14):2339-2350.e5. doi: 10.1016/j.cub.2019.06.032. Epub 2019 Jul 11. Curr Biol. 2019. PMID: 31303487 Free PMC article.
References
-
- Bellouze S, Schäfer MK, Buttigieg D, Baillat G, Rabouille C, Haase G. Golgi fragmentation in pmn mice is due to a defective ARF1/TBCE cross-talk that coordinates COPI vesicle formation and tubulin polymerization. Hum Mol Genet. 2014;23:5961–5975. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

