PaCeQuant: A Tool for High-Throughput Quantification of Pavement Cell Shape Characteristics
- PMID: 28931626
- PMCID: PMC5664455
- DOI: 10.1104/pp.17.00961
PaCeQuant: A Tool for High-Throughput Quantification of Pavement Cell Shape Characteristics
Abstract
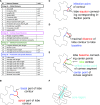
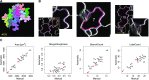
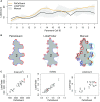
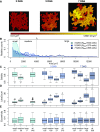
Pavement cells (PCs) are the most frequently occurring cell type in the leaf epidermis and play important roles in leaf growth and function. In many plant species, PCs form highly complex jigsaw-puzzle-shaped cells with interlocking lobes. Understanding of their development is of high interest for plant science research because of their importance for leaf growth and hence for plant fitness and crop yield. Studies of PC development, however, are limited, because robust methods are lacking that enable automatic segmentation and quantification of PC shape parameters suitable to reflect their cellular complexity. Here, we present our new ImageJ-based tool, PaCeQuant, which provides a fully automatic image analysis workflow for PC shape quantification. PaCeQuant automatically detects cell boundaries of PCs from confocal input images and enables manual correction of automatic segmentation results or direct import of manually segmented cells. PaCeQuant simultaneously extracts 27 shape features that include global, contour-based, skeleton-based, and PC-specific object descriptors. In addition, we included a method for classification and analysis of lobes at two-cell junctions and three-cell junctions, respectively. We provide an R script for graphical visualization and statistical analysis. We validated PaCeQuant by extensive comparative analysis to manual segmentation and existing quantification tools and demonstrated its usability to analyze PC shape characteristics during development and between different genotypes. PaCeQuant thus provides a platform for robust, efficient, and reproducible quantitative analysis of PC shape characteristics that can easily be applied to study PC development in large data sets.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures


Similar articles
-
User-friendly assessment of pavement cell shape features with PaCeQuant: Novel functions and tools.Methods Cell Biol. 2020;160:349-363. doi: 10.1016/bs.mcb.2020.04.010. Epub 2020 May 13. Methods Cell Biol. 2020. PMID: 32896327
-
Morphological Analysis of Leaf Epidermis Pavement Cells with PaCeQuant.Methods Mol Biol. 2019;1992:329-349. doi: 10.1007/978-1-4939-9469-4_22. Methods Mol Biol. 2019. PMID: 31148049
-
ImageJ SurfCut: a user-friendly pipeline for high-throughput extraction of cell contours from 3D image stacks.BMC Biol. 2019 May 9;17(1):38. doi: 10.1186/s12915-019-0657-1. BMC Biol. 2019. PMID: 31072374 Free PMC article.
-
Solving the Puzzle of Shape Regulation in Plant Epidermal Pavement Cells.Annu Rev Plant Biol. 2021 Jun 17;72:525-550. doi: 10.1146/annurev-arplant-080720-081920. Annu Rev Plant Biol. 2021. PMID: 34143651 Review.
-
Mechanics, geometry and genetics of epidermal cell shape regulation: different pieces of the same puzzle.Curr Opin Plant Biol. 2019 Feb;47:1-8. doi: 10.1016/j.pbi.2018.07.017. Epub 2018 Aug 28. Curr Opin Plant Biol. 2019. PMID: 30170216 Review.
Cited by
-
Mechano-chemical regulation of complex cell shape formation: Epidermal pavement cells-A case study.Quant Plant Biol. 2023 May 23;4:e5. doi: 10.1017/qpb.2023.4. eCollection 2023. Quant Plant Biol. 2023. PMID: 37251797 Free PMC article. Review.
-
MYB52 negatively regulates ADF9-meditated actin filament bundling in Arabidopsis pavement cell morphogenesis.J Integr Plant Biol. 2024 Nov;66(11):2379-2394. doi: 10.1111/jipb.13762. Epub 2024 Aug 13. J Integr Plant Biol. 2024. PMID: 39136601 Free PMC article.
-
FERONIA and microtubules independently contribute to mechanical integrity in the Arabidopsis shoot.PLoS Biol. 2021 Nov 12;19(11):e3001454. doi: 10.1371/journal.pbio.3001454. eCollection 2021 Nov. PLoS Biol. 2021. PMID: 34767544 Free PMC article.
-
ABLs and TMKs are co-receptors for extracellular auxin.Cell. 2023 Dec 7;186(25):5457-5471.e17. doi: 10.1016/j.cell.2023.10.017. Epub 2023 Nov 17. Cell. 2023. PMID: 37979582 Free PMC article.
-
Plant microphenotype: from innovative imaging to computational analysis.Plant Biotechnol J. 2024 Apr;22(4):802-818. doi: 10.1111/pbi.14244. Epub 2024 Jan 13. Plant Biotechnol J. 2024. PMID: 38217351 Free PMC article. Review.
References
-
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 - PubMed
-
- Bai Y, Falk S, Schnittger A, Jakoby MJ, Hülskamp M (2010) Tissue layer specific regulation of leaf length and width in Arabidopsis as revealed by the cell autonomous action of ANGUSTIFOLIA. Plant J 61: 191–199 - PubMed
-
- Bannigan A, Baskin TI (2005) Directional cell expansion—turning toward actin. Curr Opin Plant Biol 8: 619–624 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

