The Nitrate Transporter Family Protein LjNPF8.6 Controls the N-Fixing Nodule Activity
- PMID: 28931627
- PMCID: PMC5664486
- DOI: 10.1104/pp.17.01187
The Nitrate Transporter Family Protein LjNPF8.6 Controls the N-Fixing Nodule Activity
Abstract
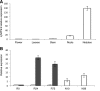
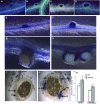
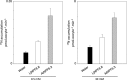
N-fixing nodules are new organs formed on legume roots as a result of the beneficial interaction with soil bacteria, rhizobia. The nodule functioning is still a poorly characterized step of the symbiotic interaction, as only a few of the genes induced in N-fixing nodules have been functionally characterized. We present here the characterization of a member of the Lotus japonicus nitrate transporter1/peptide transporter family, LjNPF8.6 The phenotypic characterization carried out in independent L. japonicus LORE1 insertion lines indicates a positive role of LjNPF8.6 on nodule functioning, as knockout mutants display N-fixation deficiency (25%) and increased nodular superoxide content. The partially compromised nodule functioning induces two striking phenotypes: anthocyanin accumulation already displayed 4 weeks after inoculation and shoot biomass deficiency, which is detected by long-term phenotyping. LjNPF8.6 achieves nitrate uptake in Xenopus laevis oocytes at both 0.5 and 30 mm external concentrations, and a possible role as a nitrate transporter in the control of N-fixing nodule activity is discussed.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures
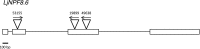




Similar articles
-
The Lotus japonicus NPF4.6 gene, encoding for a dual nitrate and ABA transporter, plays a role in the lateral root elongation process and is not involved in the N2-fixing nodule development.Plant Physiol Biochem. 2024 Nov;216:109144. doi: 10.1016/j.plaphy.2024.109144. Epub 2024 Sep 23. Plant Physiol Biochem. 2024. PMID: 39341182
-
Disruption of the Lotus japonicus transporter LjNPF2.9 increases shoot biomass and nitrate content without affecting symbiotic performances.BMC Plant Biol. 2019 Aug 30;19(1):380. doi: 10.1186/s12870-019-1978-5. BMC Plant Biol. 2019. PMID: 31470797 Free PMC article.
-
A transcription factor of the NAC family regulates nitrate-induced legume nodule senescence.New Phytol. 2023 Jun;238(5):2113-2129. doi: 10.1111/nph.18896. Epub 2023 Apr 9. New Phytol. 2023. PMID: 36945893
-
Nitrate transporters: an overview in legumes.Planta. 2017 Oct;246(4):585-595. doi: 10.1007/s00425-017-2724-6. Epub 2017 Jun 26. Planta. 2017. PMID: 28653185 Review.
-
NPF and NRT2 from Pisum sativum Potentially Involved in Nodule Functioning: Lessons from Medicago truncatula and Lotus japonicus.Plants (Basel). 2024 Jan 22;13(2):322. doi: 10.3390/plants13020322. Plants (Basel). 2024. PMID: 38276779 Free PMC article. Review.
Cited by
-
MtNPF6.5 mediates chloride uptake and nitrate preference in Medicago roots.EMBO J. 2021 Nov 2;40(21):e106847. doi: 10.15252/embj.2020106847. Epub 2021 Sep 15. EMBO J. 2021. PMID: 34523752 Free PMC article.
-
The Lotus japonicus AFB6 Gene Is Involved in the Auxin Dependent Root Developmental Program.Int J Mol Sci. 2021 Aug 6;22(16):8495. doi: 10.3390/ijms22168495. Int J Mol Sci. 2021. PMID: 34445201 Free PMC article.
-
Genome-Wide Identification and Analysis of Apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER Family (NPF) Genes Reveals MdNPF6.5 Confers High Capacity for Nitrogen Uptake under Low-Nitrogen Conditions.Int J Mol Sci. 2018 Sep 14;19(9):2761. doi: 10.3390/ijms19092761. Int J Mol Sci. 2018. PMID: 30223432 Free PMC article.
-
Organic or Inorganic Nitrogen and Rhizobia Inoculation Provide Synergistic Growth Response of a Leguminous Forb and Tree.Front Plant Sci. 2019 Oct 22;10:1308. doi: 10.3389/fpls.2019.01308. eCollection 2019. Front Plant Sci. 2019. PMID: 31695714 Free PMC article.
-
The small peptide CEP1 and the NIN-like protein NLP1 regulate NRT2.1 to mediate root nodule formation across nitrate concentrations.Plant Cell. 2023 Feb 20;35(2):776-794. doi: 10.1093/plcell/koac340. Plant Cell. 2023. PMID: 36440970 Free PMC article.
References
-
- Appleby CA. (1984) Leghemoglobin and Rhizobium respiration. Annu Rev Plant Physiol Plant Mol Biol 35: 443–478
-
- Arrese-Igor C, Gordon AJ, Minchin FR, Denison RF (1998) Nitrate entry and nitrite formation in the infected region of soybean nodules. J Exp Bot 49: 41–48
-
- Barbulova A, D’Apuzzo E, Rogato A, Chiurazzi M (2005) Improved procedures for in vitro regeneration and for phenotypical analysis in the model legume Lotus japonicus. Funct Plant Biol 32: 529–536 - PubMed
-
- Barbulova A, Rogato A, D’Apuzzo E, Omrane S, Chiurazzi M (2007) Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol Plant Microbe Interact 20: 994–1003 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

