Type B Response Regulators Act As Central Integrators in Transcriptional Control of the Auxin Biosynthesis Enzyme TAA1
- PMID: 28931628
- PMCID: PMC5664468
- DOI: 10.1104/pp.17.00878
Type B Response Regulators Act As Central Integrators in Transcriptional Control of the Auxin Biosynthesis Enzyme TAA1
Abstract
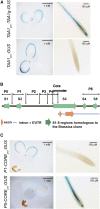
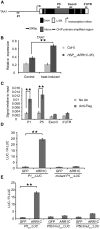
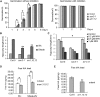
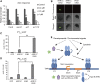
During embryogenesis and organ formation, establishing proper gradient is critical for auxin function, which is achieved through coordinated regulation of both auxin metabolism and transport. Expression of auxin biosynthetic genes is often tissue specific and is regulated by environmental signals. However, the underlying regulatory mechanisms remain elusive. Here, we investigated the transcriptional regulation of a key auxin biosynthetic gene, l-Tryptophan aminotransferase of Arabidopsis1 (TAA1). A canonical and a novel Arabidopsis (Arabidopsis thaliana) response regulator (ARR) binding site were identified in the promoter and the second intron of TAA1, which were required for its tissue-specific expression. C-termini of a subset of the type B ARRs selectively bind to one or both cis elements and activate the expression of TAA1 We further demonstrated that the ARRs not only mediate the transcriptional regulation of TAA1 by cytokinins, but also mediate its regulation by ethylene, light, and developmental signals. Through direct protein-protein interactions, the transcriptional activity of ARR1 is enhanced by ARR12, DELLAs, and ethylene-insenstive3 (EIN3). Our study thus revealed the ARR proteins act as key node that mediate the regulation of auxin biosynthesis by various hormonal, environmental, and developmental signals through transcriptional regulation of the key auxin biosynthesis gene TAA1.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures







Similar articles
-
A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis.Plant Cell. 2011 Nov;23(11):3944-60. doi: 10.1105/tpc.111.089029. Epub 2011 Nov 22. Plant Cell. 2011. PMID: 22108404 Free PMC article.
-
Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root.Plant J. 2011 May;66(3):516-27. doi: 10.1111/j.1365-313X.2011.04509.x. Epub 2011 Mar 1. Plant J. 2011. PMID: 21255165
-
TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis.Plant Cell. 2014 Jul;26(7):2889-904. doi: 10.1105/tpc.114.127993. Epub 2014 Jul 22. Plant Cell. 2014. PMID: 25052716 Free PMC article.
-
Precise Regulation of the TAA1/TAR-YUCCA Auxin Biosynthesis Pathway in Plants.Int J Mol Sci. 2023 May 10;24(10):8514. doi: 10.3390/ijms24108514. Int J Mol Sci. 2023. PMID: 37239863 Free PMC article. Review.
-
Light participates in the auxin-dependent regulation of plant growth.J Integr Plant Biol. 2021 May;63(5):819-822. doi: 10.1111/jipb.13036. Epub 2021 Feb 16. J Integr Plant Biol. 2021. PMID: 33215867 Review.
Cited by
-
The Photoperiod Stress Response in Arabidopsis thaliana Depends on Auxin Acting as an Antagonist to the Protectant Cytokinin.Int J Mol Sci. 2022 Mar 8;23(6):2936. doi: 10.3390/ijms23062936. Int J Mol Sci. 2022. PMID: 35328357 Free PMC article.
-
Shaping Ethylene Response: The Role of EIN3/EIL1 Transcription Factors.Front Plant Sci. 2019 Aug 26;10:1030. doi: 10.3389/fpls.2019.01030. eCollection 2019. Front Plant Sci. 2019. PMID: 31507622 Free PMC article. Review.
-
Cytokinin-Controlled Gradient Distribution of Auxin in Arabidopsis Root Tip.Int J Mol Sci. 2021 Apr 8;22(8):3874. doi: 10.3390/ijms22083874. Int J Mol Sci. 2021. PMID: 33918090 Free PMC article. Review.
-
Cytokinin Type-B Response Regulators Promote Bulbil Initiation in Lilium lancifolium.Int J Mol Sci. 2021 Mar 24;22(7):3320. doi: 10.3390/ijms22073320. Int J Mol Sci. 2021. PMID: 33805045 Free PMC article.
-
Response of hidden architects to salt stress.Planta. 2025 Aug 5;262(3):72. doi: 10.1007/s00425-025-04787-x. Planta. 2025. PMID: 40762673 Review.
References
-
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 - PubMed
-
- Aslanidis C, de Jong PJ, Schmitz G (1994) Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl 4: 172–177 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

