Molecular Evolution of Herpes Simplex Virus 2 Complete Genomes: Comparison between Primary and Recurrent Infections
- PMID: 28931680
- PMCID: PMC5686715
- DOI: 10.1128/JVI.00942-17
Molecular Evolution of Herpes Simplex Virus 2 Complete Genomes: Comparison between Primary and Recurrent Infections
Abstract
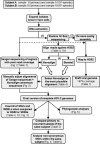
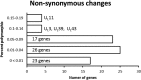
Herpes simplex virus 1 (HSV-1) and HSV-2 are large, double-stranded DNA viruses that cause lifelong persistent infections characterized by periods of quiescence and recurrent disease. How HSV evolves within an infected individual experiencing multiple episodes of recurrent disease over time is not known. We determined the genome sequences of viruses isolated from two subjects in the Herpevac Trial for Women who experienced primary HSV-2 genital disease and compared them with sequences of viruses isolated from the subsequent fifth or sixth episode of recurrent disease in the same individuals. Each of the HSV-2 genome sequences was initially obtained using next-generation sequencing and completed with Sanger sequencing. Polymorphisms over the entire genomes were mapped, and amino acid variants resulting from nonsynonymous changes were analyzed based on the secondary and tertiary structures of a previously crystallized protein. A phylogenetic reconstruction was used to assess relationships among the four HSV-2 samples, other North American sequences, and reference sequences. Little genetic drift was detected in viruses shed by the same subjects following repeated reactivation events, suggesting strong selective pressure on the viral genome to maintain sequence fidelity during reactivations from its latent state within an individual host. Our results also demonstrate that some primary HSV-2 isolates from North America more closely resemble the HG52 laboratory strain from Scotland than the low-passage-number clinical isolate SD90e from South Africa or laboratory strain 333. Thus, one of the sequences reported here would be a logical choice as a reference strain for inclusion in future studies of North American HSV-2 isolates.IMPORTANCE The extent to which the HSV-2 genome evolves during multiple episodes of reactivation from its latent state within an infected individual is not known. We used next-generation sequencing techniques to determine whole-genome sequences of four viral samples from two subjects in the Herpevac Trial. The sequence of each subject's well-documented primary isolate was compared with the sequence of the isolate from their fifth or sixth episode of recurrent disease. Only 19 genetic polymorphisms unique to the primary or recurrent isolate were identified, 10 in subject A and 9 in subject B. These observations indicate remarkable genetic conservation between primary and recurrent episodes of HSV-2 infection and imply that strong selection pressures exist to maintain the fidelity of the viral genome during repeated reactivations from its latent state. The genome conservation observed also has implications for the potential success of a therapeutic vaccine.
Keywords: Herpevac Trial for Women; complete genome; evolutionary biology; genetic polymorphisms; herpes simplex virus; next-generation sequencing.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Molecular analyses and phylogeny of the herpes simplex virus 2 US9 and glycoproteins gE/gI obtained from infected subjects during the Herpevac Trial for Women.PLoS One. 2019 Mar 8;14(3):e0212877. doi: 10.1371/journal.pone.0212877. eCollection 2019. PLoS One. 2019. PMID: 30849089 Free PMC article.
-
Genome Sequencing and Analysis of Geographically Diverse Clinical Isolates of Herpes Simplex Virus 2.J Virol. 2015 Aug;89(16):8219-32. doi: 10.1128/JVI.01303-15. Epub 2015 May 27. J Virol. 2015. PMID: 26018166 Free PMC article.
-
Genomic sequences of a low passage herpes simplex virus 2 clinical isolate and its plaque-purified derivative strain.Virology. 2014 Feb;450-451:140-5. doi: 10.1016/j.virol.2013.12.014. Epub 2013 Dec 31. Virology. 2014. PMID: 24503076 Free PMC article.
-
HSV shedding.Antiviral Res. 2004 Aug;63 Suppl 1:S19-26. doi: 10.1016/j.antiviral.2004.06.004. Antiviral Res. 2004. PMID: 15450382 Review.
-
Herpes simplex virus: the importance of asymptomatic shedding.J Antimicrob Chemother. 2000 Apr;45 Suppl T3:1-8. doi: 10.1093/jac/45.suppl_4.1. J Antimicrob Chemother. 2000. PMID: 10855766 Review.
Cited by
-
Viral Genetics Modulate Orolabial Herpes Simplex Virus Type 1 Shedding in Humans.J Infect Dis. 2019 Mar 15;219(7):1058-1066. doi: 10.1093/infdis/jiy631. J Infect Dis. 2019. PMID: 30383234 Free PMC article.
-
Genotypic and Phenotypic Diversity of Herpes Simplex Virus 2 within the Infected Neonatal Population.mSphere. 2019 Feb 27;4(1):e00590-18. doi: 10.1128/mSphere.00590-18. mSphere. 2019. PMID: 30814317 Free PMC article.
-
Herpes simplex virus 2 (HSV-2) evolves faster in cell culture than HSV-1 by generating greater genetic diversity.PLoS Pathog. 2021 Aug 26;17(8):e1009541. doi: 10.1371/journal.ppat.1009541. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34437654 Free PMC article.
-
Alphaherpesvirus Genomics: Past, Present and Future.Curr Issues Mol Biol. 2021;42:41-80. doi: 10.21775/cimb.042.041. Epub 2020 Nov 7. Curr Issues Mol Biol. 2021. PMID: 33159012 Free PMC article. Review.
-
Molecular analyses and phylogeny of the herpes simplex virus 2 US9 and glycoproteins gE/gI obtained from infected subjects during the Herpevac Trial for Women.PLoS One. 2019 Mar 8;14(3):e0212877. doi: 10.1371/journal.pone.0212877. eCollection 2019. PLoS One. 2019. PMID: 30849089 Free PMC article.
References
-
- Roizman B, Sears E. 1996. Herpes simplex viruses and their replication, p 1043–1107. In Fields BN, Knipe DM, Howley PM (ed), Fundamental virology, 3rd ed Lippincott-Raven, Philadelphia, PA.
-
- Taylor TJ, Brockman MA, McNamee EE, Knipe DM. 2002. Herpes simplex virus. Front Biosci 7:d752–d764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

