Susceptibility of Human Endogenous Retrovirus Type K to Reverse Transcriptase Inhibitors
- PMID: 28931682
- PMCID: PMC5686744
- DOI: 10.1128/JVI.01309-17
Susceptibility of Human Endogenous Retrovirus Type K to Reverse Transcriptase Inhibitors
Abstract
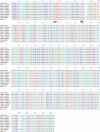
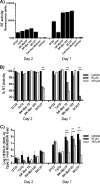
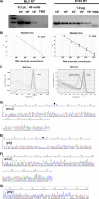
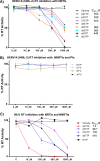
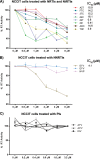
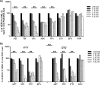
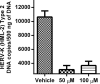
Human endogenous retroviruses (HERVs) make up 8% of the human genome. The HERV type K (HERV-K) HML-2 (HK2) family contains proviruses that are the most recent entrants into the human germ line and are transcriptionally active. In HIV-1 infection and cancer, HK2 genes produce retroviral particles that appear to be infectious, yet the replication capacity of these viruses and potential pathogenicity has been difficult to ascertain. In this report, we screened the efficacy of commercially available reverse transcriptase inhibitors (RTIs) at inhibiting the enzymatic activity of HK2 RT and HK2 genomic replication. Interestingly, only one provirus, K103, was found to encode a functional RT among those examined. Several nucleoside analogue RTIs (NRTIs) blocked K103 RT activity and consistently inhibited the replication of HK2 genomes. The NRTIs zidovudine (AZT), stavudine (d4T), didanosine (ddI), and lamivudine (3TC), and the nucleotide RTI inhibitor tenofovir (TDF), show efficacy in blocking K103 RT. HIV-1-specific nonnucleoside RTIs (NNRTIs), protease inhibitors (PIs), and integrase inhibitors (IIs) did not affect HK2, except for the NNRTI etravirine (ETV). The inhibition of HK2 infectivity by NRTIs appears to take place at either the reverse transcription step of the viral genome prior to HK2 viral particle formation and/or in the infected cells. Inhibition of HK2 by these drugs will be useful in suppressing HK2 infectivity if these viruses prove to be pathogenic in cancer, neurological disorders, or other diseases associated with HK2. The present studies also elucidate a key aspect of the life cycle of HK2, specifically addressing how they do, and/or did, replicate.IMPORTANCE Endogenous retroviruses are relics of ancestral virus infections in the human genome. The most recent of these infections was caused by HK2. While HK2 often remains silent in the genome, this group of viruses is activated in HIV-1-infected and cancer cells. Recent evidence suggests that these viruses are infectious, and the potential exists for HK2 to contribute to disease. We show that HK2, and specifically the enzyme that mediates virus replication, can be inhibited by a panel of drugs that are commercially available. We show that several drugs block HK2 with different efficacies. The inhibition of HK2 replication by antiretroviral drugs appears to occur in the virus itself as well as after infection of cells. Therefore, these drugs might prove to be an effective treatment by suppressing HK2 infectivity in diseases where these viruses have been implicated, such as cancer and neurological syndromes.
Keywords: AZT; HERV-K; NNRTIs; NRTIs; NtRTIs; reverse transcriptase inhibitors.
Copyright © 2017 American Society for Microbiology.
Figures
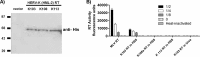
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

