Longitudinal analysis of colon crypt stem cell dynamics in sulindac treated Familial Adenomatous Polyposis patients
- PMID: 28931879
- PMCID: PMC5607292
- DOI: 10.1038/s41598-017-11865-y
Longitudinal analysis of colon crypt stem cell dynamics in sulindac treated Familial Adenomatous Polyposis patients
Abstract
The non-steroidal anti-inflammatory drug sulindac decreases size and number of adenomas after 4-6 months of treatment for familial adenomatous polyposis (FAP) patients. However, the underlying mechanism remains unknown. As stem cells are thought to be the tumor precursor cells, visualizing their behavior is crucial for monitoring tumor progression. Increased tag diversity in inactive genes is indicative of a protracted clonal evolution and consequently, increased risk for tumor formation. Therefore, the effect of sulindac on stem cell dynamics was studied. Normal appearing single crypts were laser microdissected in placebo- and sulindac- treated FAP patient tissue after which the methylation patterns were visualized by Next Generation Sequencing. A significant difference in tag diversity over time was found in the sulindac group compared to the placebo group (*p = 0.018), indicative of a shortened clonal evolution treated sulindac. The rate of change in tag diversity over time was correlated with polyp number change over time. No significant difference over time was observed in the percent methylation when comparing placebo vs sulindac. In conclusion, daily sulindac administration in FAP patients significantly altered colorectal stem cell dynamics, which might explain the chemopreventive action of this drug indicating that tag diversity may be used as a predictive biomarker.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
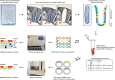
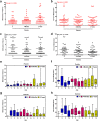
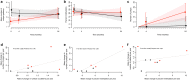
References
-
- Wright NA, Irwin M. The kinetics of villus cell populations in the mouse small intestine. I. Normal villi: the steady state requirement. Cell Tissue Kinet. 1982;15:595–609. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

