Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2
- PMID: 28931898
- PMCID: PMC5607329
- DOI: 10.1038/s41598-017-12212-x
Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2
Abstract
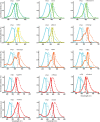
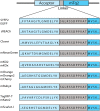
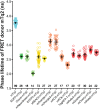
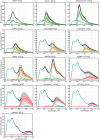
The performance of Förster Resonance Energy Transfer (FRET) biosensors depends on brightness and photostability, which are dependent on the characteristics of the fluorescent proteins that are employed. Yellow fluorescent protein (YFP) is often used as an acceptor but YFP is prone to photobleaching and pH changes. In this study, we evaluated the properties of a diverse set of acceptor fluorescent proteins in combination with the optimized CFP variant mTurquoise2 as the donor. To determine the theoretical performance of acceptors, the Förster radius was determined. The practical performance was determined by measuring FRET efficiency and photostability of tandem fusion proteins in mammalian cells. Our results show that mNeonGreen is the most efficient acceptor for mTurquoise2 and that the photostability is better than SYFP2. The non-fluorescent YFP variant sREACh is an efficient acceptor, which is useful in lifetime-based FRET experiments. Among the orange and red fluorescent proteins, mCherry and mScarlet-I are the best performing acceptors. Several new pairs were applied in a multimolecular FRET based sensor for detecting activation of a heterotrimeric G-protein by G-protein coupled receptors. Overall, the sensor with mNeonGreen as acceptor and mTurquoise2 as donor showed the highest dynamic range in ratiometric FRET imaging experiments with the G-protein sensor.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Tsien RY. The Green Fluorescent. Proteins. 1998;67:509–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

