Strain Level Streptococcus Colonization Patterns during the First Year of Life
- PMID: 28932211
- PMCID: PMC5592222
- DOI: 10.3389/fmicb.2017.01661
Strain Level Streptococcus Colonization Patterns during the First Year of Life
Abstract
Pneumococcal pneumonia has decreased significantly since the implementation of the pneumococcal conjugate vaccine (PCV), nevertheless, in many developing countries pneumonia mortality in infants remains high. We have undertaken a study of the nasopharyngeal (NP) microbiome during the first year of life in infants from The Philippines and South Africa. The study entailed the determination of the Streptococcus sp. carriage using a lytA qPCR assay, whole metagenomic sequencing, and in silico serotyping of Streptococcus pneumoniae, as well as 16S rRNA amplicon based community profiling. The lytA carriage in both populations increased with infant age and lytA+ samples ranged from 24 to 85% of the samples at each sampling time point. We next developed informatic tools for determining Streptococcus community composition and pneumococcal serotype from metagenomic sequences derived from a subset of longitudinal lytA-positive Streptococcus enrichment cultures from The Philippines (n = 26 infants, 50% vaccinated) and South African (n = 7 infants, 100% vaccinated). NP samples from infants were passaged in enrichment media, and metagenomic DNA was purified and sequenced. In silico capsular serotyping of these 51 metagenomic assemblies assigned known serotypes in 28 samples, and the co-occurrence of serotypes in 5 samples. Eighteen samples were not typeable using known serotypes but did encode for capsule biosynthetic cluster genes similar to non-encapsulated reference sequences. In addition, we performed metagenomic assembly and 16S rRNA amplicon profiling to understand co-colonization dynamics of Streptococcus sp. and other NP genera, revealing the presence of multiple Streptococcus species as well as potential respiratory pathogens in healthy infants. A range of virulence and drug resistant elements were identified as circulating in the NP microbiomes of these infants. This study revealed the frequent co-occurrence of multiple S. pneumoniae strains along with Streptococcus sp. and other potential pathogens such as S. aureus in the NP microbiome of these infants. In addition, the in silico serotype analysis proved powerful in determining the serotypes in S. pneumoniae carriage, and may lead to developing better targeted vaccines to prevent invasive pneumococcal disease (IPD) in these countries. These findings suggest that NP colonization by S. pneumoniae during the first years of life is a dynamic process involving multiple serotypes and species.
Keywords: Serotypes; Streptococcus pneumoniae; nasopharyngeal microbiome; pneumococcal conjugate vaccine.
Figures
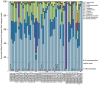
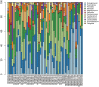
References
-
- Adegbola R. A., DeAntonio R., Hill P. C., Roca A., Usuf E., Hoet B., et al. . (2014). Carriage of Streptococcus Pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PLoS ONE 9:e103293. 10.1371/journal.pone.0103293 - DOI - PMC - PubMed
-
- Blumental S., Granger-Farbos A., Moisi J. C., Soullie B., Leroy P., Njanpop-Lafourcade B. M., et al. . (2015). Virulence factors of Streptococcus Pneumoniae. comparison between African and French invasive isolates and implication for future vaccines. PLoS ONE 10:e0133885. 10.1371/journal.pone.0133885 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

