Shrimp miR-10a Is Co-opted by White Spot Syndrome Virus to Increase Viral Gene Expression and Viral Replication
- PMID: 28932224
- PMCID: PMC5592198
- DOI: 10.3389/fimmu.2017.01084
Shrimp miR-10a Is Co-opted by White Spot Syndrome Virus to Increase Viral Gene Expression and Viral Replication
Abstract
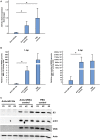
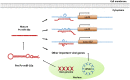
Members of the microRNA miR-10 family are highly conserved and play many important roles in diverse biological mechanisms, including immune-related responses and cancer-related processes in certain types of cancer. In this study, we found the most highly upregulated shrimp microRNA from Penaeus vannamei during white spot syndrome virus (WSSV) infection was miR-10a. After confirming the expression level of miR-10a by northern blot and quantitative RT-PCR, an in vivo experiment showed that the viral copy number was decreased in miR-10a-inhibited shrimp. We found that miR-10a targeted the 5' untranslated region (UTR) of at least three viral genes (vp26, vp28, and wssv102), and plasmids that were controlled by the 5' UTR of these genes produced enhanced luciferase signals in transfected SF9 cells. These results suggest a previously unreported role for shrimp miR-10a and even a new type of host-virus interaction, whereby a co-opts the key cellular regulator miR-10a to globally enhance the translation of viral proteins.
Keywords: Penaeus vannamei; miR-10a; microRNA; virus–host interaction; white spot syndrome virus.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

