The anti-SLAMF7 antibody elotuzumab mediates NK cell activation through both CD16-dependent and -independent mechanisms
- PMID: 28932638
- PMCID: PMC5599094
- DOI: 10.1080/2162402X.2017.1339853
The anti-SLAMF7 antibody elotuzumab mediates NK cell activation through both CD16-dependent and -independent mechanisms
Abstract
Elotuzumab is a humanized therapeutic monoclonal antibody directed to the surface glycoprotein SLAMF7 (CS1, CRACC, CD319), which is highly expressed on multiple myeloma (MM) tumor cells. Improved clinical outcomes have been observed following treatment of MM patients with elotuzumab in combination with lenalidomide or bortezomib. Previous work showed that elotuzumab stimulates NK cell-mediated antibody-dependent cellular cytotoxicity (ADCC), via Fc-domain engagement with FcγRIIIa (CD16). SLAMF7 is also expressed on NK cells, where it can transmit stimulatory signals. We tested whether elotuzumab can directly activate NK cells via ligation with SLAMF7 on NK cells in addition to targeting ADCC through CD16. We show that elotuzumab strongly promoted degranulation and activation of NK cells in a CD16-dependent manner, and a non-fucosylated form of elotuzumab with higher affinity to CD16 exhibited enhanced potency. Using F(ab')2 or Fc-mutant forms of the antibody, the direct binding of elotuzumab to SLAMF7 alone could not stimulate measurable CD69 expression or degranulation of NK cells. However, the addition of soluble elotuzumab could costimulate calcium signaling responses triggered by multimeric engagement of NKp46 and NKG2D in a CD16-independent manner. Thus, while elotuzumab primarily stimulates NK cells through CD16, it can also transduce effective "trans"-costimulatory signals upon direct engagement with SLAMF7, since these responses did not require direct co-engagement with the activating receptors. Trans-costimulation by elotuzumab has potential to reduce activation thresholds of other NK cell receptors engaging with their ligands on myeloma target cell surfaces, thereby potentially further increasing NK cell responsiveness in patients.
Keywords: ADCC; NK cells; SLAMF7; elotuzumab; multiple myeloma.
Figures
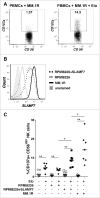
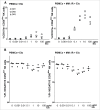
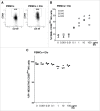
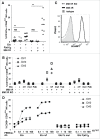
References
-
- Palumbo A, Anderson K. Multiple myeloma. N Eng J Med 2011; 364:1046-60; PMID:21410373; https://doi.org/10.1056/NEJMra1011442 - DOI - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66:7-30 - PubMed
-
- Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol 2012; 9:135-43; PMID:22349016; https://doi.org/10.1038/nrclinonc.2012.15 - DOI - PubMed
-
- Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol 2010; 10:317-27; PMID:20414205; https://doi.org/10.1038/nri2744 - DOI - PMC - PubMed
-
- Palumbo A, Sonneveld P. Preclinical and clinical evaluation of elotuzumab, a SLAMF7-targeted humanized monoclonal antibody in development for multiple myeloma. Expert Rev Hematol 2015; 8(4):481-91:1-11 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
