The Gut Commensal Microbiome of Drosophila melanogaster Is Modified by the Endosymbiont Wolbachia
- PMID: 28932814
- PMCID: PMC5597968
- DOI: 10.1128/mSphere.00287-17
The Gut Commensal Microbiome of Drosophila melanogaster Is Modified by the Endosymbiont Wolbachia
Abstract
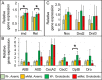
Endosymbiotic Wolbachia bacteria and the gut microbiome have independently been shown to affect several aspects of insect biology, including reproduction, development, life span, stem cell activity, and resistance to human pathogens, in insect vectors. This work shows that Wolbachia bacteria, which reside mainly in the fly germline, affect the microbial species present in the fly gut in a lab-reared strain. Drosophila melanogaster hosts two main genera of commensal bacteria-Acetobacter and Lactobacillus. Wolbachia-infected flies have significantly reduced titers of Acetobacter. Sampling of the microbiome of axenic flies fed with equal proportions of both bacteria shows that the presence of Wolbachia bacteria is a significant determinant of the composition of the microbiome throughout fly development. However, this effect is host genotype dependent. To investigate the mechanism of microbiome modulation, the effect of Wolbachia bacteria on Imd and reactive oxygen species pathways, the main regulators of immune response in the fly gut, was measured. The presence of Wolbachia bacteria does not induce significant changes in the expression of the genes for the effector molecules in either pathway. Furthermore, microbiome modulation is not due to direct interaction between Wolbachia bacteria and gut microbes. Confocal analysis shows that Wolbachia bacteria are absent from the gut lumen. These results indicate that the mechanistic basis of the modulation of composition of the microbiome by Wolbachia bacteria is more complex than a direct bacterial interaction or the effect of Wolbachia bacteria on fly immunity. The findings reported here highlight the importance of considering the composition of the gut microbiome and host genetic background during Wolbachia-induced phenotypic studies and when formulating microbe-based disease vector control strategies. IMPORTANCEWolbachia bacteria are intracellular bacteria present in the microbiome of a large fraction of insects and parasitic nematodes. They can block mosquitos' ability to transmit several infectious disease-causing pathogens, including Zika, dengue, chikungunya, and West Nile viruses and malaria parasites. Certain extracellular bacteria present in the gut lumen of these insects can also block pathogen transmission. However, our understanding of interactions between Wolbachia and gut bacteria and how they influence each other is limited. Here we show that the presence of Wolbachia strain wMel changes the composition of gut commensal bacteria in the fruit fly. Our findings implicate interactions between bacterial species as a key factor in determining the overall composition of the microbiome and thus reveal new paradigms to consider in the development of disease control strategies.
Keywords: Drosophila microbiome; Wolbachia; gut microbiome; symbiosis.
Figures

Similar articles
-
Wolbachia endosymbiotic bacteria alter the gut microbiome in the fly Drosophila nigrosparsa.J Invertebr Pathol. 2023 Jun;198:107915. doi: 10.1016/j.jip.2023.107915. Epub 2023 Mar 21. J Invertebr Pathol. 2023. PMID: 36958642
-
Intracellular Density of Wolbachia Is Mediated by Host Autophagy and the Bacterial Cytoplasmic Incompatibility Gene cifB in a Cell Type-Dependent Manner in Drosophila melanogaster.mBio. 2021 Jan 12;12(1):e02205-20. doi: 10.1128/mBio.02205-20. mBio. 2021. PMID: 33436431 Free PMC article.
-
Effects of co-occurring Wolbachia and Spiroplasma endosymbionts on the Drosophila immune response against insect pathogenic and non-pathogenic bacteria.BMC Microbiol. 2016 Feb 9;16:16. doi: 10.1186/s12866-016-0634-6. BMC Microbiol. 2016. PMID: 26862076 Free PMC article.
-
Tephritid-microbial interactions to enhance fruit fly performance in sterile insect technique programs.BMC Microbiol. 2019 Dec 24;19(Suppl 1):287. doi: 10.1186/s12866-019-1650-0. BMC Microbiol. 2019. PMID: 31870316 Free PMC article. Review.
-
Wolbachia endosymbionts and human disease control.Mol Biochem Parasitol. 2014 Jul;195(2):88-95. doi: 10.1016/j.molbiopara.2014.07.004. Epub 2014 Jul 18. Mol Biochem Parasitol. 2014. PMID: 25046729 Review.
Cited by
-
The Role of Microbiota in Drosophila melanogaster Aging.Front Aging. 2022 May 19;3:909509. doi: 10.3389/fragi.2022.909509. eCollection 2022. Front Aging. 2022. PMID: 35821860 Free PMC article. Review.
-
Ixodes scapularis does not harbor a stable midgut microbiome.ISME J. 2018 Nov;12(11):2596-2607. doi: 10.1038/s41396-018-0161-6. Epub 2018 Jun 26. ISME J. 2018. PMID: 29946195 Free PMC article.
-
Alzheimer's disease and gut-brain axis: Drosophila melanogaster as a model.Front Neurosci. 2025 Feb 4;19:1543826. doi: 10.3389/fnins.2025.1543826. eCollection 2025. Front Neurosci. 2025. PMID: 39967802 Free PMC article. Review.
-
Pervasive tissue-, genetic background-, and allele-specific gene expression effects in Drosophila melanogaster.PLoS Genet. 2024 Aug 23;20(8):e1011257. doi: 10.1371/journal.pgen.1011257. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39178312 Free PMC article.
-
Bacterial recognition by PGRP-SA and downstream signalling by Toll/DIF sustain commensal gut bacteria in Drosophila.PLoS Genet. 2022 Jan 10;18(1):e1009992. doi: 10.1371/journal.pgen.1009992. eCollection 2022 Jan. PLoS Genet. 2022. PMID: 35007276 Free PMC article.
References
-
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi:10.1073/pnas.1218525110. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

