Learning directed acyclic graphs from large-scale genomics data
- PMID: 28933027
- PMCID: PMC5607220
- DOI: 10.1186/s13637-017-0063-3
Learning directed acyclic graphs from large-scale genomics data
Abstract
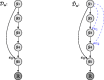
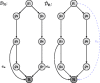
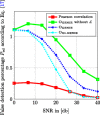
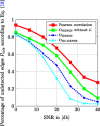
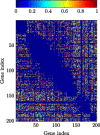
In this paper, we consider the problem of learning the genetic interaction map, i.e., the topology of a directed acyclic graph (DAG) of genetic interactions from noisy double-knockout (DK) data. Based on a set of well-established biological interaction models, we detect and classify the interactions between genes. We propose a novel linear integer optimization program called the Genetic-Interactions-Detector (GENIE) to identify the complex biological dependencies among genes and to compute the DAG topology that matches the DK measurements best. Furthermore, we extend the GENIE program by incorporating genetic interaction profile (GI-profile) data to further enhance the detection performance. In addition, we propose a sequential scalability technique for large sets of genes under study, in order to provide statistically significant results for real measurement data. Finally, we show via numeric simulations that the GENIE program and the GI-profile data extended GENIE (GI-GENIE) program clearly outperform the conventional techniques and present real data results for our proposed sequential scalability technique.
Keywords: Big data; Discrete optimization; Genetic interaction analysis; Graph learning; Large-scale gene networks; Multiple hypothesis test.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

