Human Coronaviruses: A Review of Virus-Host Interactions
- PMID: 28933406
- PMCID: PMC5456285
- DOI: 10.3390/diseases4030026
Human Coronaviruses: A Review of Virus-Host Interactions
Abstract
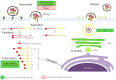
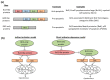
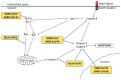
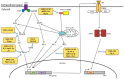
Human coronaviruses (HCoVs) are known respiratory pathogens associated with a range of respiratory outcomes. In the past 14 years, the onset of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) have thrust HCoVs into spotlight of the research community due to their high pathogenicity in humans. The study of HCoV-host interactions has contributed extensively to our understanding of HCoV pathogenesis. In this review, we discuss some of the recent findings of host cell factors that might be exploited by HCoVs to facilitate their own replication cycle. We also discuss various cellular processes, such as apoptosis, innate immunity, ER stress response, mitogen-activated protein kinase (MAPK) pathway and nuclear factor kappa B (NF-κB) pathway that may be modulated by HCoVs.
Keywords: ER stress; MAPK; NF-κB; apoptosis; human coronavirus; innate immunity; virus–host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Vijgen L., Keyaerts E., Moës E., Maes P., Duson G., van Ranst M. Development of One-Step, Real-Time, Quantitative Reverse Transcriptase PCR Assays for Absolute Quantitation of Human Coronaviruses OC43 and 229E. J. Clin. Microbiol. 2005;43:5452–5456. doi: 10.1128/JCM.43.11.5452-5456.2005. - DOI - PMC - PubMed
-
- Jones B.A., Grace D., Kock R., Alonso S., Rushton J., Said M.Y., McKeever D., Mutua F., Young J., McDermott J., et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. USA. 2013;21:8399–8340. doi: 10.1073/pnas.1208059110. - DOI - PMC - PubMed
-
- Van der Hoek L. Human coronaviruses: What do they cause? Antivir. Ther. 2007;12:651–658. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

