The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance
- PMID: 28933623
- PMCID: PMC5788422
- DOI: 10.1080/15592294.2017.1381807
The defining DNA methylation signature of Kabuki syndrome enables functional assessment of genetic variants of unknown clinical significance
Abstract
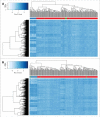
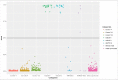
Kabuki syndrome (KS) is caused by mutations in KMT2D, which is a histone methyltransferase involved in methylation of H3K4, a histone marker associated with DNA methylation. Analysis of >450,000 CpGs in 24 KS patients with pathogenic mutations in KMT2D and 216 controls, identified 24 genomic regions, along with 1,504 CpG sites with significant DNA methylation changes including a number of Hox genes and the MYO1F gene. Using the most differentiating and significant probes and regions we developed a "methylation variant pathogenicity (MVP) score," which enables 100% sensitive and specific identification of individuals with KS, which was confirmed using multiple public and internal patient DNA methylation databases. We also demonstrated the ability of the MVP score to accurately reclassify variants of unknown significance in subjects with apparent clinical features of KS, enabling its potential use in molecular diagnostics. These findings provide novel insights into the molecular etiology of KS and illustrate that DNA methylation patterns can be interpreted as 'epigenetic echoes' in certain clinical disorders.
Keywords: DNA methylation; KDM6A; KMT2D; Kabuki syndrome; variant classification.
Figures


References
-
- Bögershausen N, Gatinois V, Riehmer V, Kayserili H, Becker J, Thoenes M, Simsek‐Kiper PÖ, Barat‐Houari M, Elcioglu NH, Wieczorek D, Tinschert S. Mutation Update for Kabuki Syndrome Genes KMT2D and KDM6A and Further Delineation of X‐Linked Kabuki Syndrome Subtype 2. Hum Mutat. 2016;37(9): 847-64. doi:10.1002/humu.23026. PMID:27302555 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
