acn-1, a C. elegans homologue of ACE, genetically interacts with the let-7 microRNA and other heterochronic genes
- PMID: 28933985
- PMCID: PMC5628652
- DOI: 10.1080/15384101.2017.1344798
acn-1, a C. elegans homologue of ACE, genetically interacts with the let-7 microRNA and other heterochronic genes
Abstract
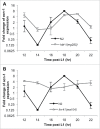
The heterochronic pathway in C. elegans controls the relative timing of cell fate decisions during post-embryonic development. It includes a network of microRNAs (miRNAs), such as let-7, and protein-coding genes, such as the stemness factors, LIN-28 and LIN-41. Here we identified the acn-1 gene, a homologue of mammalian angiotensin-converting enzyme (ACE), as a new suppressor of the stem cell developmental defects of let-7 mutants. Since acn-1 null mutants die during early larval development, we used RNAi to characterize the role of acn-1 in C. elegans seam cell development, and determined its interaction with heterochronic factors, including let-7 and its downstream interactors - lin-41, hbl-1, and apl-1. We demonstrate that although RNAi knockdown of acn-1 is insufficient to cause heterochronic defects on its own, loss of acn-1 suppresses the retarded phenotypes of let-7 mutants and enhances the precocious phenotypes of hbl-1, though not lin-41, mutants. Conversely, the pattern of acn-1 expression, which oscillates during larval development, is disrupted by lin-41 mutants but not by hbl-1 mutants. Finally, we show that acn-1(RNAi) enhances the let-7-suppressing phenotypes caused by loss of apl-1, a homologue of the Alzheimer's disease-causing amyloid precursor protein (APP), while significantly disrupting the expression of apl-1 during the L4 larval stage. In conclusion, acn-1 interacts with heterochronic genes and appears to function downstream of let-7 and its target genes, including lin-41 and apl-1.
Keywords: APP; acn-1; developmental timing; heterochronic genes; let-7; lin-41.
Figures



References
-
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol [Internet]. 2002;18:495-513. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12142272. doi: 10.1146/annurev.cellbio.18.012502.105832 - DOI - PubMed
-
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol [Internet]. 2007;17:R425-34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17550772. doi: 10.1016/j.cub.2007.03.043 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
