Applications of PNA-Based Artificial Restriction DNA Cutters
- PMID: 28934140
- PMCID: PMC6151779
- DOI: 10.3390/molecules22101586
Applications of PNA-Based Artificial Restriction DNA Cutters
Abstract
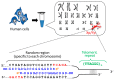
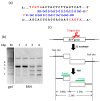
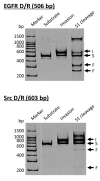
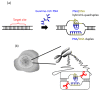
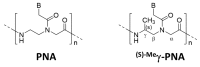
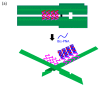
More than ten years ago, artificial restriction DNA cutters were developed by combining two pseudo-complementary peptide nucleic acid (pcPNA) strands with either Ce(IV)/EDTA or S1 nuclease. They have remarkably high site-specificity and can cut only one predetermined site in the human genome. In this article, recent progress of these man-made tools have been reviewed. By cutting the human genome site-selectively, desired fragments can be clipped from either the termini of chromosomes (telomeres) or from the middle of genome. These fragments should provide important information on the biological functions of complicated genome system. DNA/RNA hybrid duplexes, which are formed in living cells, are also site-selectively hydrolyzed by these cutters. In order to further facilitate the applications of the artificial DNA cutters, various chemical modifications have been attempted. One of the most important successes is preparation of PNA derivatives which can form double-duplex invasion complex even under high salt conditions. This is important for in vivo applications, since the inside of living cells is abundant of metal ions. Furthermore, site-selective DNA cutters which require only one PNA strand, in place of a pair of pcPNA strands, are developed. This progress has opened a way to new fields of PNA-based biochemistry and biotechnology.
Keywords: DNA/RNA hybrid; PNA invasion; site-selective DNA cutter; site-selective genome scission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Origin of high fidelity in target-sequence recognition by PNA-Ce(IV)/EDTA combinations as site-selective DNA cutters.J Am Chem Soc. 2009 Feb 25;131(7):2657-62. doi: 10.1021/ja808290e. J Am Chem Soc. 2009. PMID: 19199631
-
Chemical modifications of artificial restriction DNA cutter (ARCUT) to promote its in vivo and in vitro applications.Artif DNA PNA XNA. 2014 Dec 15;5(3):e1112457. doi: 10.1080/1949095X.2015.1112457. Artif DNA PNA XNA. 2014. PMID: 26744220 Free PMC article. Review.
-
Site-selective scission of human genome using PNA-based artificial restriction DNA cutter.Methods Mol Biol. 2014;1050:111-20. doi: 10.1007/978-1-62703-553-8_9. Methods Mol Biol. 2014. PMID: 24297354
-
PNA-NLS conjugates as single-molecular activators of target sites in double-stranded DNA for site-selective scission.Org Biomol Chem. 2013 Aug 28;11(32):5233-8. doi: 10.1039/c3ob40947c. Org Biomol Chem. 2013. PMID: 23820872
-
Artificial DNA cutters for DNA manipulation and genome engineering.Chem Soc Rev. 2011 Dec;40(12):5657-68. doi: 10.1039/c1cs15039a. Epub 2011 May 12. Chem Soc Rev. 2011. PMID: 21566825 Review.
Cited by
-
Pseudo-Complementary G:C Base Pair for Mixed Sequence dsDNA Invasion and Its Applications in Diagnostics (SARS-CoV-2 Detection).JACS Au. 2023 Feb 1;3(2):449-458. doi: 10.1021/jacsau.2c00588. eCollection 2023 Feb 27. JACS Au. 2023. PMID: 36873687 Free PMC article.
-
Special Issue: Molecular Properties and the Applications of Peptide Nucleic Acids.Molecules. 2018 Aug 8;23(8):1977. doi: 10.3390/molecules23081977. Molecules. 2018. PMID: 30096770 Free PMC article.
-
Enzymatic Synthesis of Chemical Nuclease Triplex-Forming Oligonucleotides with Gene-Silencing Applications.Nucleic Acids Res. 2022 Jun 10;50(10):5467-5481. doi: 10.1093/nar/gkac438. Nucleic Acids Res. 2022. PMID: 35640595 Free PMC article.
-
Phosphodiester models for cleavage of nucleic acids.Beilstein J Org Chem. 2018 Apr 10;14:803-837. doi: 10.3762/bjoc.14.68. eCollection 2018. Beilstein J Org Chem. 2018. PMID: 29719577 Free PMC article. Review.
-
The shaping of a molecular linguist: How a career studying DNA energetics revealed the language of molecular communication.J Biol Chem. 2021 Jan-Jun;296:100522. doi: 10.1016/j.jbc.2021.100522. Epub 2021 Apr 7. J Biol Chem. 2021. PMID: 34237886 Free PMC article.
References
-
- Komiyama M., Yoshimoto K., Sisido M., Ariga K. Chemistry can make strict and fuzzy controls for bio-systems: DNA nanoarchitectonics and cell-macromolecular nanoarchitectonics. Bull. Chem. Soc. Jpn. 2017;90:967–1004. doi: 10.1246/bcsj.20170156. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

