Denitrification potential of the eastern oyster microbiome using a 16S rRNA gene based metabolic inference approach
- PMID: 28934286
- PMCID: PMC5608302
- DOI: 10.1371/journal.pone.0185071
Denitrification potential of the eastern oyster microbiome using a 16S rRNA gene based metabolic inference approach
Abstract
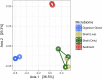
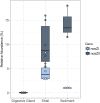
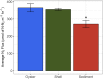
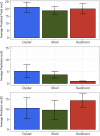
The eastern oyster (Crassostrea virginica) is a foundation species providing significant ecosystem services. However, the roles of oyster microbiomes have not been integrated into any of the services, particularly nitrogen removal through denitrification. We investigated the composition and denitrification potential of oyster microbiomes with an approach that combined 16S rRNA gene analysis, metabolic inference, qPCR of the nitrous oxide reductase gene (nosZ), and N2 flux measurements. Microbiomes of the oyster digestive gland, the oyster shell, and sediments adjacent to the oyster reef were examined based on next generation sequencing (NGS) of 16S rRNA gene amplicons. Denitrification potentials of the microbiomes were determined by metabolic inferences using a customized denitrification gene and genome database with the paprica (PAthway PRediction by phylogenetIC plAcement) bioinformatics pipeline. Denitrification genes examined included nitrite reductase (nirS and nirK) and nitrous oxide reductase (nosZ), which was further subdivided by genotype into clade I (nosZI) or clade II (nosZII). Continuous flow through experiments measuring N2 fluxes were conducted with the oysters, shells, and sediments to compare denitrification activities. Paprica properly classified the composition of microbiomes, showing similar classification results from Silva, Greengenes and RDP databases. Microbiomes of the oyster digestive glands and shells were quite different from each other and from the sediments. The relative abundance of denitrifying bacteria inferred by paprica was higher in oysters and shells than in sediments suggesting that oysters act as hotspots for denitrification in the marine environment. Similarly, the inferred nosZI gene abundances were also higher in the oyster and shell microbiomes than in the sediment microbiome. Gene abundances for nosZI were verified with qPCR of nosZI genes, which showed a significant positive correlation (F1,7 = 14.7, p = 6.0x10-3, R2 = 0.68). N2 flux rates were significantly higher in the oyster (364.4 ± 23.5 μmol N-N2 m-2 h-1) and oyster shell (355.3 ± 6.4 μmol N-N2 m-2 h-1) compared to the sediment (270.5 ± 20.1 μmol N-N2 m-2 h-1). Thus, bacteria carrying nosZI genes were found to be an important denitrifier, facilitating nitrogen removal in oyster reefs. In addition, this is the first study to validate the use of 16S gene based metabolic inference as a method for determining microbiome function, such as denitrification, by comparing inference results with qPCR gene quantification and rate measurements.
Conflict of interest statement
Figures



Similar articles
-
Nitrogen removal and spatial distribution of denitrifier and anammox communities in a bioreactor for mine drainage treatment.Water Res. 2014 Dec 1;66:350-360. doi: 10.1016/j.watres.2014.08.038. Epub 2014 Sep 4. Water Res. 2014. PMID: 25233117
-
Microbiome Analysis Reveals Diversity and Function of Mollicutes Associated with the Eastern Oyster, Crassostrea virginica.mSphere. 2021 May 12;6(3):e00227-21. doi: 10.1128/mSphere.00227-21. mSphere. 2021. PMID: 33980678 Free PMC article.
-
NosZI microbial community determined the potential of denitrification and nitrous oxide emission in river sediments of Qinghai-Tibetan Plateau.Environ Res. 2022 Nov;214(Pt 4):114138. doi: 10.1016/j.envres.2022.114138. Epub 2022 Aug 18. Environ Res. 2022. PMID: 35988830
-
Beyond Bioextraction: The Role of Oyster-Mediated Denitrification in Nutrient Management.Environ Sci Technol. 2021 Nov 2;55(21):14457-14465. doi: 10.1021/acs.est.1c01901. Epub 2021 Oct 21. Environ Sci Technol. 2021. PMID: 34672569 Free PMC article. Review.
-
Oyster disease in a changing environment: Decrypting the link between pathogen, microbiome and environment.Mar Environ Res. 2019 Jan;143:124-140. doi: 10.1016/j.marenvres.2018.11.007. Epub 2018 Nov 16. Mar Environ Res. 2019. PMID: 30482397 Review.
Cited by
-
Host Species and Environment Shape the Gut Microbiota of Cohabiting Marine Bivalves.Microb Ecol. 2023 Oct;86(3):1755-1772. doi: 10.1007/s00248-023-02192-z. Epub 2023 Feb 22. Microb Ecol. 2023. PMID: 36811710 Free PMC article.
-
Persistent tissue-specific resident microbiota in oysters across a broad geographical range.Environ Microbiol Rep. 2024 Oct;16(5):e70026. doi: 10.1111/1758-2229.70026. Environ Microbiol Rep. 2024. PMID: 39446070 Free PMC article.
-
Microbiome differences in sugarcane and metabolically engineered oilcane accessions and their implications for bioenergy production.Biotechnol Biofuels Bioprod. 2023 Mar 30;16(1):56. doi: 10.1186/s13068-023-02302-6. Biotechnol Biofuels Bioprod. 2023. PMID: 36998044 Free PMC article.
-
Comparative Genomic Analysis of Three Pseudomonas Species Isolated from the Eastern Oyster (Crassostrea virginica) Tissues, Mantle Fluid, and the Overlying Estuarine Water Column.Microorganisms. 2021 Feb 27;9(3):490. doi: 10.3390/microorganisms9030490. Microorganisms. 2021. PMID: 33673397 Free PMC article.
-
Bacterial metagenome analysis of Mytilus galloprovincialis collected from Istanbul and Izmir coastal stations of Turkey.Environ Monit Assess. 2020 Feb 19;192(3):186. doi: 10.1007/s10661-020-8129-1. Environ Monit Assess. 2020. PMID: 32072329
References
-
- Glibert PM, Hinkle DC, Sturgis B, Jesien RV. Eutrophication of a Maryland/Virginia coastal lagoon: a tipping point, ecosystem changes, and potential causes. Estuaries and Coasts. 2014;37(S1):128–46. doi: 10.1007/s12237-013-9630-3 - DOI
-
- Cerco CF, Noel MR. Can oyster restoration reverse cultural eutrophication in Chesapeake Bay? Estuaries and Coasts. 2007;30(2):331–43.
-
- Newell RIE, Cornwell JC, Owens MS. Influence of simulated bivalve biodeposition and microphytobenthos on sediment nitrogen dynamics: A laboratory study. Limnol Oceanogr. 2002;47(5):1367–79.
-
- Kellogg ML, Smyth AR, Luckenbach MW, Carmichael RH, Brown BL, Cornwell JC, et al. Use of oysters to mitigate eutrophication in coastal waters. Estuar Coast Shelf Sci. 2014;151:156–68.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

