hnRNP L controls HPV16 RNA polyadenylation and splicing in an Akt kinase-dependent manner
- PMID: 28934469
- PMCID: PMC5766200
- DOI: 10.1093/nar/gkx606
hnRNP L controls HPV16 RNA polyadenylation and splicing in an Akt kinase-dependent manner
Abstract
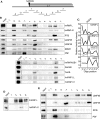
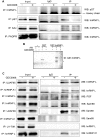
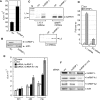
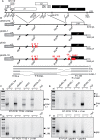
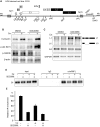
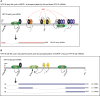
Inhibition of the Akt kinase activates HPV16 late gene expression by reducing HPV16 early polyadenylation and by activating HPV16 late L1 mRNA splicing. We identified 'hot spots' for RNA binding proteins at the early polyA signal and at splice sites on HPV16 late mRNAs. We observed that hnRNP L was associated with sequences at all HPV16 late splice sites and at the early polyA signal. Akt kinase inhibition resulted in hnRNP L dephosphorylation and reduced association of hnRNP L with HPV16 mRNAs. This was accompanied by an increased binding of U2AF65 and Sam68 to HPV16 mRNAs. Furthermore, siRNA knock-down of hnRNP L or Akt induced HPV16 gene expression. Treatment of HPV16 immortalized keratinocytes with Akt kinase inhibitor reduced hnRNP L binding to HPV16 mRNAs and induced HPV16 L1 mRNA production. Finally, deletion of the hnRNP L binding sites in HPV16 subgenomic expression plasmids resulted in activation of HPV16 late gene expression. In conclusion, the Akt kinase inhibits HPV16 late gene expression at the level of RNA processing by controlling the RNA-binding protein hnRNP L. We speculate that Akt kinase activity upholds an intracellular milieu that favours HPV16 early gene expression and suppresses HPV16 late gene expression.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Chow L.T., Broker T.R., Steinberg B.M.. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010; 118:422–449. - PubMed
-
- Schiffman M., Doorbar J., Wentzensen N., de Sanjose S., Fakhry C., Monk B.J., Stanley M.A., Franceschi S.. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers. 2016; 2:16086. - PubMed
-
- Mighty K.K., Laimins L.A.. The role of human papillomaviruses in oncogenesis. Rec. Results Cancer Res. 2014; 193:135–148. - PubMed
-
- Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N.. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999; 189:12–19. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

