The conserved AU dinucleotide at the 5' end of nascent U1 snRNA is optimized for the interaction with nuclear cap-binding-complex
- PMID: 28934473
- PMCID: PMC5766165
- DOI: 10.1093/nar/gkx608
The conserved AU dinucleotide at the 5' end of nascent U1 snRNA is optimized for the interaction with nuclear cap-binding-complex
Abstract
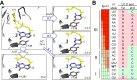
Splicing is initiated by a productive interaction between the pre-mRNA and the U1 snRNP, in which a short RNA duplex is established between the 5' splice site of a pre-mRNA and the 5' end of the U1 snRNA. A long-standing puzzle has been why the AU dincucleotide at the 5'-end of the U1 snRNA is highly conserved, despite the absence of an apparent role in the formation of the duplex. To explore this conundrum, we varied this AU dinucleotide into all possible permutations and analyzed the resulting molecular consequences. This led to the unexpected findings that the AU dinucleotide dictates the optimal binding of cap-binding complex (CBC) to the 5' end of the nascent U1 snRNA, which ultimately influences the utilization of U1 snRNP in splicing. Our data also provide a structural interpretation as to why the AU dinucleotide is conserved during evolution.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Luhrmann R.. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009; 36:593–608. - PubMed
-
- Will C.L., Luhrmann R.. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 1997; 9:320–328. - PubMed
-
- Chang T.H., Tung L., Yeh F.L., Chen J.H., Chang S.L.. Functions of the DExD/H-box proteins in nuclear pre-mRNA splicing. Biochim. Biophys. Acta. 2013; 1829:764–774. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

