Human DNA polymerase delta double-mutant D316A;E318A interferes with DNA mismatch repair in vitro
- PMID: 28934474
- PMCID: PMC5766205
- DOI: 10.1093/nar/gkx611
Human DNA polymerase delta double-mutant D316A;E318A interferes with DNA mismatch repair in vitro
Abstract
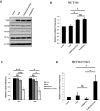
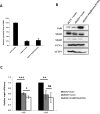
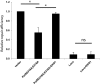
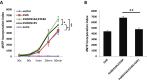
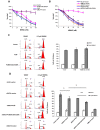
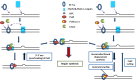
DNA mismatch repair (MMR) is a highly-conserved DNA repair mechanism, whose primary role is to remove DNA replication errors preventing them from manifesting as mutations, thereby increasing the overall genome stability. Defects in MMR are associated with increased cancer risk in humans and other organisms. Here, we characterize the interaction between MMR and a proofreading-deficient allele of the human replicative DNA polymerase delta, PolδD316A;E318A, which has a higher capacity for strand displacement DNA synthesis than wild type Polδ. Human cell lines overexpressing PolδD316A;E318A display a mild mutator phenotype, while nuclear extracts of these cells exhibit reduced MMR activity in vitro, and these defects are complemented by overexpression or addition of exogenous human Exonuclease 1 (EXO1). By contrast, another proofreading-deficient mutant, PolδD515V, which has a weaker strand displacement activity, does not decrease the MMR activity as significantly as PolδD316A;E318A. In addition, PolδD515V does not increase the mutation frequency in MMR-proficient cells. Based on our findings, we propose that the proofreading activity restricts the strand displacement activity of Polδ in MMR. This contributes to maintain the nicks required for EXO1 entry, and in this manner ensures the dominance of the EXO1-dependent MMR pathway.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Modrich P., Lahue R.. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996; 65:101–133. - PubMed
-
- Li G.M. Mechanisms and functions of DNA mismatch repair. Cell research. 2008; 18:85–98. - PubMed
-
- Jascur T., Boland C.R.. Structure and function of the components of the human DNA mismatch repair system. Int. J. Cancer. 2006; 119:2030–2035. - PubMed
-
- Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996; 10:1433–1442. - PubMed
-
- Schofield M.J., Hsieh P.. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003; 57:579–608. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

