De novo pathway-based biomarker identification
- PMID: 28934488
- PMCID: PMC5766193
- DOI: 10.1093/nar/gkx642
De novo pathway-based biomarker identification
Abstract
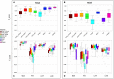
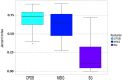
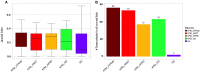
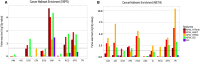
Gene expression profiles have been extensively discussed as an aid to guide the therapy by predicting disease outcome for the patients suffering from complex diseases, such as cancer. However, prediction models built upon single-gene (SG) features show poor stability and performance on independent datasets. Attempts to mitigate these drawbacks have led to the development of network-based approaches that integrate pathway information to produce meta-gene (MG) features. Also, MG approaches have only dealt with the two-class problem of good versus poor outcome prediction. Stratifying patients based on their molecular subtypes can provide a detailed view of the disease and lead to more personalized therapies. We propose and discuss a novel MG approach based on de novo pathways, which for the first time have been used as features in a multi-class setting to predict cancer subtypes. Comprehensive evaluation in a large cohort of breast cancer samples from The Cancer Genome Atlas (TCGA) revealed that MGs are considerably more stable than SG models, while also providing valuable insight into the cancer hallmarks that drive them. In addition, when tested on an independent benchmark non-TCGA dataset, MG features consistently outperformed SG models. We provide an easy-to-use web service at http://pathclass.compbio.sdu.dk where users can upload their own gene expression datasets from breast cancer studies and obtain the subtype predictions from all the classifiers.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Arijs I., Quintens R., Van Lommel L., Van Steen K., De Hertogh G., Lemaire K., Schraenen A., Perrier C., Van Assche G., Vermeire S. et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm. Bowel Dis. 2010; 16:2090–2098. - PubMed
-
- van ’t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415:530–536. - PubMed
-
- Karnes R.J., Bergstralh E.J., Davicioni E., Ghadessi M., Buerki C., Mitra A.P., Crisan A., Erho N., Vergara I.A., Lam L.L. et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J. Urol. 2013; 190:2047–2053. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

