Number of inadvertent RNA targets for morpholino knockdown in Danio rerio is largely underestimated: evidence from the study of Ser/Arg-rich splicing factors
- PMID: 28934490
- PMCID: PMC5766196
- DOI: 10.1093/nar/gkx638
Number of inadvertent RNA targets for morpholino knockdown in Danio rerio is largely underestimated: evidence from the study of Ser/Arg-rich splicing factors
Abstract
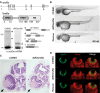
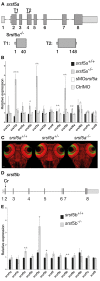
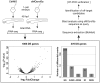
Although the involvement of Ser/Arg-rich (SR) proteins in RNA metabolism is well documented, their role in vertebrate development remains elusive. We, therefore, elected to take advantage of the zebrafish model organism to study the SR genes' functions using the splicing morpholino (sMO) microinjection and the programmable site-specific nucleases. Consistent with previous research, we revealed discrepancies between the mutant and morphant phenotypes and we show that these inconsistencies may result from a large number of unsuspected inadvertent morpholino RNA targets. While microinjection of MOs directed against srsf5a (sMOsrsf5a) led to developmental defects, the corresponding homozygous mutants did not display any phenotypic traits. Furthermore, microinjection of sMOsrsf5a into srsf5a-/- led to the previously observed morphant phenotype. Similar findings were observed for other SR genes. sMOsrsf5a alternative target genes were identified using deep mRNA sequencing. We uncovered that only 11 consecutive bases complementary to sMOsrsf5a are sufficient for binding and subsequent blocking of splice sites. In addition, we observed that sMOsrsf5a secondary targets can be reduced by increasing embryos growth temperature after microinjection. Our data contribute to the debate about MO specificity, efficacy and the number of unknown targeted sequences.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Deletion of morpholino binding sites (DeMOBS) to assess specificity of morphant phenotypes.Sci Rep. 2020 Sep 21;10(1):15366. doi: 10.1038/s41598-020-71708-1. Sci Rep. 2020. PMID: 32958829 Free PMC article.
-
Design and Microinjection of Morpholino Antisense Oligonucleotides and mRNA into Zebrafish Embryos to Elucidate Specific Gene Function in Heart Development.J Vis Exp. 2022 Aug 9;(186):10.3791/63324. doi: 10.3791/63324. J Vis Exp. 2022. PMID: 36036621 Free PMC article.
-
Application of Zebrafish and Knockdown Technology to Define Progranulin Neuronal Function.Methods Mol Biol. 2018;1806:207-231. doi: 10.1007/978-1-4939-8559-3_15. Methods Mol Biol. 2018. PMID: 29956279
-
A primer for morpholino use in zebrafish.Zebrafish. 2009 Mar;6(1):69-77. doi: 10.1089/zeb.2008.0555. Zebrafish. 2009. PMID: 19374550 Free PMC article. Review.
-
Knockout, Knockdown, and the Schrödinger Paradox: Genetic Immunity to Phenotypic Recapitulation in Zebrafish.Genes (Basel). 2024 Sep 3;15(9):1164. doi: 10.3390/genes15091164. Genes (Basel). 2024. PMID: 39336755 Free PMC article. Review.
Cited by
-
Differential Alternative Splicing Genes and Isoform Regulation Networks of Rapeseed (Brassica napus L.) Infected with Sclerotinia sclerotiorum.Genes (Basel). 2020 Jul 13;11(7):784. doi: 10.3390/genes11070784. Genes (Basel). 2020. PMID: 32668742 Free PMC article.
-
CRISPR-RfxCas13d screening uncovers Bckdk as a post-translational regulator of the maternal-to-zygotic transition in teleosts.bioRxiv [Preprint]. 2024 May 23:2024.05.22.595167. doi: 10.1101/2024.05.22.595167. bioRxiv. 2024. PMID: 38826327 Free PMC article. Preprint.
-
Versatile Genome Engineering Techniques Advance Human Ocular Disease Researches in Zebrafish.Front Cell Dev Biol. 2018 Jul 12;6:75. doi: 10.3389/fcell.2018.00075. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 30050903 Free PMC article. Review.
-
Differential alternative splicing genes and isoform co-expression networks of Brassica napus under multiple abiotic stresses.Front Plant Sci. 2022 Oct 13;13:1009998. doi: 10.3389/fpls.2022.1009998. eCollection 2022. Front Plant Sci. 2022. PMID: 36311064 Free PMC article.
-
Casting CRISPR-Cas13d to fish for microprotein functions in animal development.iScience. 2022 Nov 10;25(12):105547. doi: 10.1016/j.isci.2022.105547. eCollection 2022 Dec 22. iScience. 2022. PMID: 36444300 Free PMC article. Review.
References
-
- Long J.C., Caceres J.F.. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009; 417:15–27. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

