Phosphorylation regulates human polη stability and damage bypass throughout the cell cycle
- PMID: 28934491
- PMCID: PMC5766190
- DOI: 10.1093/nar/gkx619
Phosphorylation regulates human polη stability and damage bypass throughout the cell cycle
Abstract
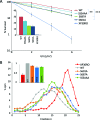
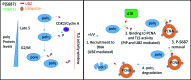
DNA translesion synthesis (TLS) is a crucial damage tolerance pathway that oversees the completion of DNA replication in the presence of DNA damage. TLS polymerases are capable of bypassing a distorted template but they are generally considered inaccurate and they need to be tightly regulated. We have previously shown that polη is phosphorylated on Serine 601 after DNA damage and we have demonstrated that this modification is important for efficient damage bypass. Here we report that polη is also phosphorylated by CDK2, in the absence of damage, in a cell cycle-dependent manner and we identify serine 687 as an important residue targeted by the kinase. We discover that phosphorylation on serine 687 regulates the stability of the polymerase during the cell cycle, allowing it to accumulate in late S and G2 when productive TLS is critical for cell survival. Furthermore, we show that alongside the phosphorylation of S601, the phosphorylation of S687 and S510, S512 and/or S514 are important for damage bypass and cell survival after UV irradiation. Taken together our results provide new insights into how cells can, at different times, modulate DNA TLS for improved cell survival.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Lehmann A.R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J.F., Kannouche P.L., Green C.M.. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst). 2007; 6:891–899. - PubMed
-
- Bienko M., Green C.M., Sabbioneda S., Crosetto N., Matic I., Hibbert R.G., Begovic T., Niimi A., Mann M., Lehmann A.R. et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol. Cell. 2010; 37:396–407. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

