Derivatives of Bst-like Gss-polymerase with improved processivity and inhibitor tolerance
- PMID: 28934494
- PMCID: PMC5766155
- DOI: 10.1093/nar/gkx645
Derivatives of Bst-like Gss-polymerase with improved processivity and inhibitor tolerance
Abstract
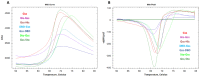
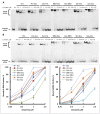
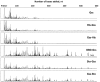
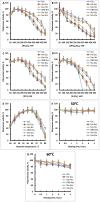
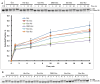
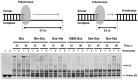
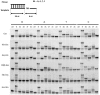
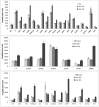
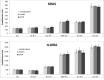
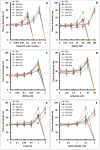
At the moment, one of the actual trends in medical diagnostics is a development of methods for practical applications such as point-of-care testing, POCT or research tools, for example, whole genome amplification, WGA. All the techniques are based on using of specific DNA polymerases having strand displacement activity, high synthetic processivity, fidelity and, most significantly, tolerance to contaminants, appearing from analysed biological samples or collected under purification procedures. Here, we have designed a set of fusion enzymes based on catalytic domain of DNA polymerase I from Geobacillus sp. 777 with DNA-binding domain of DNA ligase Pyrococcus abyssi and Sto7d protein from Sulfolobus tokodaii, analogue of Sso7d. Designed chimeric DNA polymerases DBD-Gss, Sto-Gss and Gss-Sto exhibited the same level of thermal stability, thermal transferase activity and fidelity as native Gss; however, the processivity was increased up to 3-fold, leading to about 4-fold of DNA product in WGA which is much more exiting. The attachment of DNA-binding proteins enhanced the inhibitor tolerance of chimeric polymerases in loop-mediated isothermal amplification to several of the most common DNA sample contaminants-urea and whole blood, heparin, ethylenediaminetetraacetic acid, NaCl, ethanol. Therefore, chimeric Bst-like Gss-polymerase will be promising tool for both WGA and POCT due to increased processivity and inhibitor tolerance.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Burgers P.M., Koonin E.V., Bruford E., Blanco L., Burtis K.C., Christman M.F., Copeland W.C., Friedberg E.C., Hanaoka F., Hinkle D.C. et al. . Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001; 276:43487–43490. - PubMed
-
- Hübscher U., Spadari S., Villani G., Maga G.. DNA Polymerases Discovery, Characterization and Functions in Cellular DNA Transactions. 2010; Singapore: World Scientific Publishing Co. Pte. Ltd.
-
- Patel P.H., Suzuki M., Adman E., Shinkai a, Loeb L.A.. Prokaryotic DNA polymerase I: evolution, structure, and ‘base flipping’ mechanism for nucleotide selection. J. Mol. Biol. 2001; 308:823–837. - PubMed
-
- Joyce C.M., Kelley W.S., Grindley N.D.. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J. Biol. Chem. 1982; 257:1958–1964. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

