Combining asymmetric 13C-labeling and isotopic filter/edit NOESY: a novel strategy for rapid and logical RNA resonance assignment
- PMID: 28934505
- PMCID: PMC5766159
- DOI: 10.1093/nar/gkx591
Combining asymmetric 13C-labeling and isotopic filter/edit NOESY: a novel strategy for rapid and logical RNA resonance assignment
Abstract
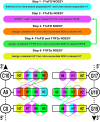
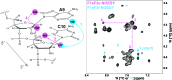
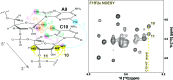
Although ∼98% of the human genomic output is transcribed as non-protein coding RNA, <2% of the protein data bank structures comprise RNA. This huge structural disparity stems from combined difficulties of crystallizing RNA for X-ray crystallography along with extensive chemical shift overlap and broadened linewidths associated with NMR of RNA. While half of the deposited RNA structures in the PDB were solved by NMR methods, the usefulness of NMR is still limited by the high cost of sample preparation and challenges of resonance assignment. Here we propose a novel strategy for resonance assignment that combines new strategic 13C labeling technologies with filter/edit type NOESY experiments to greatly reduce spectral complexity and crowding. This new strategy allowed us to assign important non-exchangeable resonances of proton and carbon (1', 2', 2, 5, 6 and 8) nuclei using only one sample and <24 h of NMR instrument time for a 27 nt model RNA. The method was further extended to assigning a 6 nt bulge from a 61 nt viral RNA element justifying its use for a wide range RNA chemical shift resonance assignment problems.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
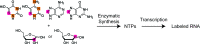




References
-
- Mattick J.S. A new paradigm for developmental biology. J. Exp. Biol. 2007; 210:1526–1547. - PubMed
-
- Steitz T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008; 9:242–253. - PubMed
-
- Ramakrishnan V. The ribosome emerges from a black box. Cell. 2014; 159:979–984. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

