A comprehensive, cell specific microRNA catalogue of human peripheral blood
- PMID: 28934507
- PMCID: PMC5766192
- DOI: 10.1093/nar/gkx706
A comprehensive, cell specific microRNA catalogue of human peripheral blood
Abstract
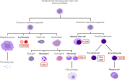
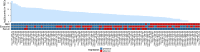
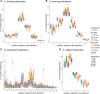
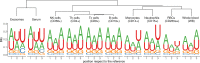
With this study, we provide a comprehensive reference dataset of detailed miRNA expression profiles from seven types of human peripheral blood cells (NK cells, B lymphocytes, cytotoxic T lymphocytes, T helper cells, monocytes, neutrophils and erythrocytes), serum, exosomes and whole blood. The peripheral blood cells from buffy coats were typed and sorted using FACS/MACS. The overall dataset was generated from 450 small RNA libraries using high-throughput sequencing. By employing a comprehensive bioinformatics and statistical analysis, we show that 3' trimming modifications as well as composition of 3' added non-templated nucleotides are distributed in a lineage-specific manner-the closer the hematopoietic progenitors are, the higher their similarities in sequence variation of the 3' end. Furthermore, we define the blood cell-specific miRNA and isomiR expression patterns and identify novel cell type specific miRNA candidates. The study provides the most comprehensive contribution to date towards a complete miRNA catalogue of human peripheral blood, which can be used as a reference for future studies. The dataset has been deposited in GEO and also can be explored interactively following this link: http://134.245.63.235/ikmb-tools/bloodmiRs.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. - PubMed
-
- Lewis B.P., Burge C.B., Bartel D.P.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120:15–20. - PubMed
-
- Iorio M. V., Ferracin M., Liu C.-G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. et al. . MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005; 65:7065–7070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

