Identification of novel candidate disease genes from de novo exonic copy number variants
- PMID: 28934986
- PMCID: PMC5607840
- DOI: 10.1186/s13073-017-0472-7
Identification of novel candidate disease genes from de novo exonic copy number variants
Abstract
Background: Exon-targeted microarrays can detect small (<1000 bp) intragenic copy number variants (CNVs), including those that affect only a single exon. This genome-wide high-sensitivity approach increases the molecular diagnosis for conditions with known disease-associated genes, enables better genotype-phenotype correlations, and facilitates variant allele detection allowing novel disease gene discovery.
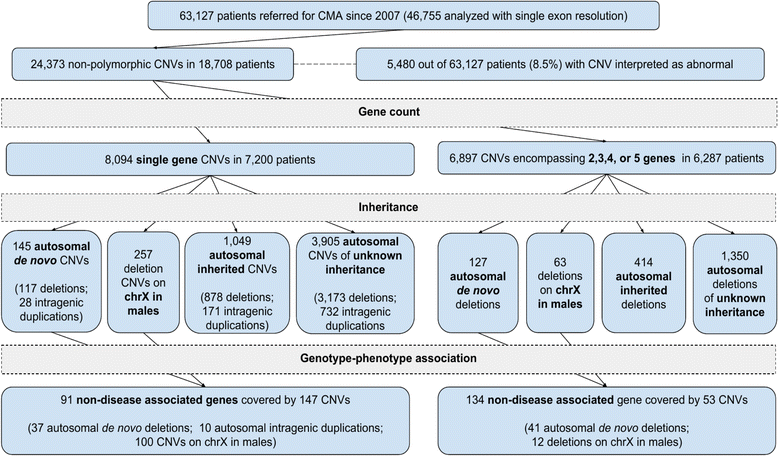
Methods: We retrospectively analyzed data from 63,127 patients referred for clinical chromosomal microarray analysis (CMA) at Baylor Genetics laboratories, including 46,755 individuals tested using exon-targeted arrays, from 2007 to 2017. Small CNVs harboring a single gene or two to five non-disease-associated genes were identified; the genes involved were evaluated for a potential disease association.
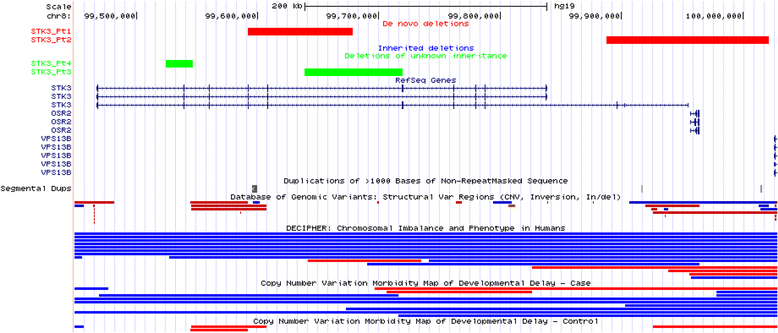
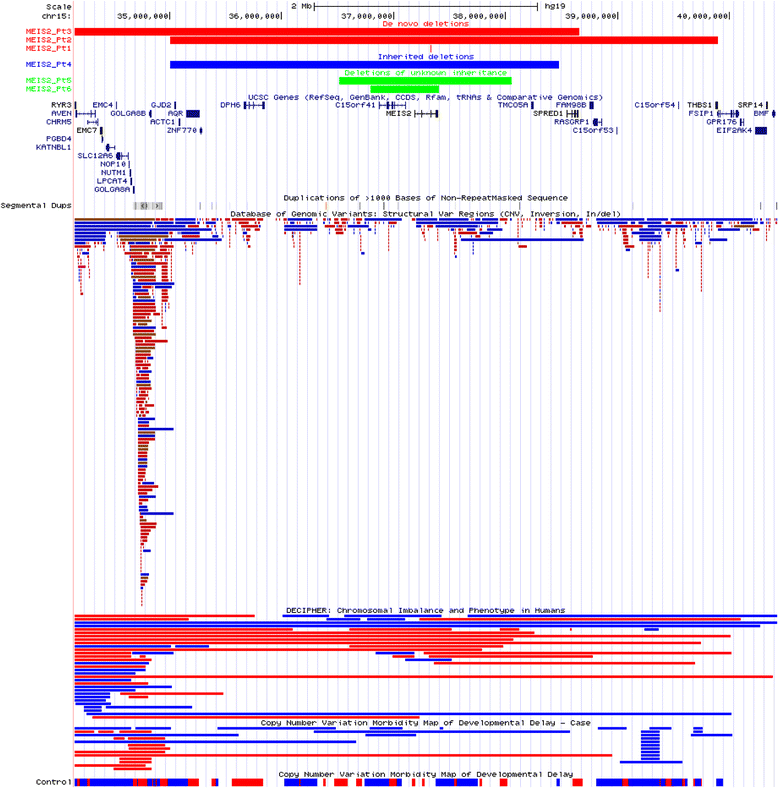
Results: In this clinical population, among rare CNVs involving any single gene reported in 7200 patients (11%), we identified 145 de novo autosomal CNVs (117 losses and 28 intragenic gains), 257 X-linked deletion CNVs in males, and 1049 inherited autosomal CNVs (878 losses and 171 intragenic gains); 111 known disease genes were potentially disrupted by de novo autosomal or X-linked (in males) single-gene CNVs. Ninety-one genes, either recently proposed as candidate disease genes or not yet associated with diseases, were disrupted by 147 single-gene CNVs, including 37 de novo deletions and ten de novo intragenic duplications on autosomes and 100 X-linked CNVs in males. Clinical features in individuals with de novo or X-linked CNVs encompassing at most five genes (224 bp to 1.6 Mb in size) were compared to those in individuals with larger-sized deletions (up to 5 Mb in size) in the internal CMA database or loss-of-function single nucleotide variants (SNVs) detected by clinical or research whole-exome sequencing (WES). This enabled the identification of recently published genes (BPTF, NONO, PSMD12, TANGO2, and TRIP12), novel candidate disease genes (ARGLU1 and STK3), and further confirmation of disease association for two recently proposed disease genes (MEIS2 and PTCHD1). Notably, exon-targeted CMA detected several pathogenic single-exon CNVs missed by clinical WES analyses.
Conclusions: Together, these data document the efficacy of exon-targeted CMA for detection of genic and exonic CNVs, complementing and extending WES in clinical diagnostics, and the potential for discovery of novel disease genes by genome-wide assay.
Keywords: CNVs; Exon targeted array CGH; Intragenic copy number variants; de novo variants.
Conflict of interest statement
Ethics approval and consent to participate
Approval for this study, including written informed consents (H-22769) and waivers of informed consent (H-37568 and H-36612), were obtained from the Baylor College of Medicine Institutional Review Board. The Baylor College of Medicine IRB (IORG number 0000055) is recognized by the United States Office of Human Research Protections (OHRP) and Food and Drug Administration (FDA) under the federal-wide assurance program. The Baylor College of Medicine IRB is also fully accredited by the Association for the Accreditation of Human Research Protection Programs (AAHRPP).
Consent for publication
The Editor has waived consent to publish the clinical information in the manuscript due to the minimal risk of identification.
Competing interests
BCM and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), formerly the Baylor Miraca Genetics Laboratories (BMGL), which performs chromosomal microarray analysis and clinical exome sequencing. PL, CAB, WB, JAR, SRL, MW, YY, AMB, JLS, SWC, AP, CAS, and PS are employees of BCM and derive support through a professional services agreement with the BG. JRL serves on the Scientific Advisory Board of the BG. JRL has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

