Toward the identification of a type I toxin-antitoxin system in the plasmid DNA of dairy Lactobacillus rhamnosus
- PMID: 28935987
- PMCID: PMC5608710
- DOI: 10.1038/s41598-017-12218-5
Toward the identification of a type I toxin-antitoxin system in the plasmid DNA of dairy Lactobacillus rhamnosus
Abstract
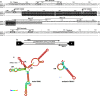
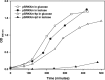
Plasmids carry genes that give bacteria beneficial traits and allow them to survive in competitive environments. In many cases, they also harbor toxin-antitoxin (TA) systems necessary for plasmid maintenance. TA systems are generally characterized by a stable "toxin", a protein or peptide capable of killing the cell upon plasmid loss and by an unstable "antitoxin", a protein or a non-coding RNA that inhibits toxin activity. Here we report data toward the identification of a RNA-regulated TA system in the plasmid DNA of L. rhamnosus isolated from cheese. The proposed TA system comprises two convergently transcribed RNAs: a toxin RNA encoding a 29 amino acid peptide named Lpt and an antitoxin non-coding RNA. Both toxin and antitoxin RNAs resulted upregulated under conditions mimicking cheese ripening. The toxicity of the Lpt peptide was demonstrated in E. coli by cloning the Lpt ORF under the control of an inducible promoter. Bioinformatics screening of the bacterial nucleotide database, shows that regions homologous to the Lpt TA locus are widely distributed in the Lactobacillus genus, particularly within the L. casei group, suggesting a relevant role of TA systems in plasmid maintenance of cheese microbiota.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
TasA-tasB, a new putative toxin-antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE-doc TA system.BMC Genomics. 2006 Oct 13;7:259. doi: 10.1186/1471-2164-7-259. BMC Genomics. 2006. PMID: 17038198 Free PMC article.
-
Functional characterization of the type I toxin Lpt from Lactobacillus rhamnosus by fluorescence and atomic force microscopy.Sci Rep. 2019 Oct 23;9(1):15208. doi: 10.1038/s41598-019-51523-z. Sci Rep. 2019. PMID: 31645607 Free PMC article.
-
Shared mechanisms of enhanced plasmid maintenance and antibiotic tolerance mediated by the VapBC toxin:antitoxin system.mBio. 2025 Feb 5;16(2):e0261624. doi: 10.1128/mbio.02616-24. Epub 2024 Dec 20. mBio. 2025. PMID: 39704502 Free PMC article.
-
Type I toxin-antitoxin systems in bacteria: from regulation to biological functions.EcoSal Plus. 2024 Dec 12;12(1):eesp00252022. doi: 10.1128/ecosalplus.esp-0025-2022. Epub 2024 May 20. EcoSal Plus. 2024. PMID: 38767346 Free PMC article. Review.
-
Type I Toxin-Antitoxin Systems: Regulating Toxin Expression via Shine-Dalgarno Sequence Sequestration and Small RNA Binding.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0030-2018. doi: 10.1128/microbiolspec.RWR-0030-2018. Microbiol Spectr. 2018. PMID: 30051800 Free PMC article. Review.
Cited by
-
Stopping Untreatable Pathogen Infections Using Peptide Ligands to Sabotage Pathogenic Cell Surface Proteins.Mol Biotechnol. 2019 Aug;61(8):602-609. doi: 10.1007/s12033-019-00189-z. Mol Biotechnol. 2019. PMID: 31161299 Review.
-
Expression of DinJ-YafQ System of Lactobacillus casei Group Strains in Response to Food Processing Stresses.Microorganisms. 2019 Oct 11;7(10):438. doi: 10.3390/microorganisms7100438. Microorganisms. 2019. PMID: 31614503 Free PMC article.
-
Microbiota of Cheese Ecosystems: A Perspective on Cheesemaking.Foods. 2025 Feb 27;14(5):830. doi: 10.3390/foods14050830. Foods. 2025. PMID: 40077532 Free PMC article. Review.
-
Strategies to Investigate Membrane Damage, Nucleoid Condensation, and RNase Activity of Bacterial Toxin-Antitoxin Systems.Methods Protoc. 2021 Oct 8;4(4):71. doi: 10.3390/mps4040071. Methods Protoc. 2021. PMID: 34698227 Free PMC article.
-
Bacterial Type I Toxins: Folding and Membrane Interactions.Toxins (Basel). 2021 Jul 14;13(7):490. doi: 10.3390/toxins13070490. Toxins (Basel). 2021. PMID: 34357962 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

