Analytical Validation and Capabilities of the Epic CTC Platform: Enrichment-Free Circulating Tumour Cell Detection and Characterization
- PMID: 28936239
- PMCID: PMC5572988
- DOI: 10.5772/60725
Analytical Validation and Capabilities of the Epic CTC Platform: Enrichment-Free Circulating Tumour Cell Detection and Characterization
Abstract
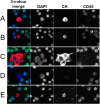
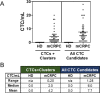
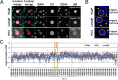
The Epic Platform was developed for the unbiased detection and molecular characterization of circulating tumour cells (CTCs). Here, we report assay performance data, including accuracy, linearity, specificity and intra/inter-assay precision of CTC enumeration in healthy donor (HD) blood samples spiked with varying concentrations of cancer cell line controls (CLCs). Additionally, we demonstrate clinical feasibility for CTC detection in a small cohort of metastatic castrate-resistant prostate cancer (mCRPC) patients. The Epic Platform demonstrated accuracy, linearity and sensitivity for the enumeration of all CLC concentrations tested. Furthermore, we established the precision between multiple operators and slide staining batches and assay specificity showing zero CTCs detected in 18 healthy donor samples. In a clinical feasibility study, at least one traditional CTC/mL (CK+, CD45-, and intact nuclei) was detected in 89 % of 44 mCRPC samples, whereas 100 % of samples had CTCs enumerated if additional CTC subpopulations (CK-/CD45- and CK+ apoptotic CTCs) were included in the analysis. In addition to presenting Epic Platform's performance with respect to CTC enumeration, we provide examples of its integrated downstream capabilities, including protein biomarker expression and downstream genomic analyses at single cell resolution.
Keywords: Analytical Validation; Biomarker; CTC; CTM; Circulating Tumour Cells; Clinical Feasibility; Epic CTC Platform; Fluid Biopsy; Liquid Biopsy; Metastasis.
Figures


References
-
- Gupta G P and Massague J (2006) Cancer metastasis: Building a framework. Cell. 127: 679–95 - PubMed
-
- Chaffer C L and Weinberg R A (2011) A perspective on cancer cell metastasis. Science. 331: 1559–64 - PubMed
-
- Hanahan D and Weinberg R A (2011) Hallmarks of cancer: The next generation. Cell. 144: 646–74 - PubMed
-
- Yu M Bardia A Wittner B S Stott S L Smas M E Ting D T Isakoff S J Ciciliano J C Wells M N Shah A M Concannon K F Donaldson M C Sequist L V Brachtel E Sgroi D Baselga J Ramaswamy S Toner M Haber D A and Maheswaran S (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 339: 580–4 - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

