Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a
- PMID: 28937654
- PMCID: PMC5666722
- DOI: 10.3390/ijms18102040
Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a
Abstract
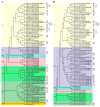
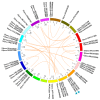
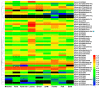
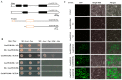
Plant CIRCADIAN CLOCK ASSOCIATED1 (CCA1)-like proteins are a class of single-repeat MYELOBLASTOSIS ONCOGENE (MYB) transcription factors generally featured by a highly conserved motif SHAQK(Y/F)F, which play important roles in multiple biological processes. Soybean is an important grain legume for seed protein and edible vegetable oil. However, essential understandings regarding CCA1-like proteins are very limited in soybean. In this study, 54 CCA1-like proteins were identified by data mining of soybean genome. Phylogenetic analysis indicated that soybean CCA1-like subfamily showed evolutionary conservation and diversification. These CCA1-like genes displayed tissue-specific expression patterns, and analysis of genomic organization and evolution revealed 23 duplicated gene pairs. Among them, GmMYB138a was chosen for further investigation. Our protein-protein interaction studies revealed that GmMYB138a, but not its alternatively spliced isoform, interacts with a 14-3-3 protein (GmSGF14l). Although GmMYB138a was predominately localized in nucleus, the resulting complex of GmMYB138a and GmSGF14l was almost evenly distributed in nucleus and cytoplasm, supporting that 14-3-3s interact with their clients to alter their subcellular localization. Additionally, qPCR analysis suggested that GmMYB138a and GmSGF14l synergistically or antagonistically respond to drought, cold and salt stresses. Our findings will contribute to future research in regard to functions of soybean CCA1-like subfamily, especially regulatory mechanisms of GmMYB138a in response to abiotic stresses.
Keywords: 14-3-3; CCA1-like proteins; MYB; protein interaction; soybean.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Soybean CCA1-like MYB transcription factor GmMYB133 modulates isoflavonoid biosynthesis.Biochem Biophys Res Commun. 2018 Dec 9;507(1-4):324-329. doi: 10.1016/j.bbrc.2018.11.033. Epub 2018 Nov 15. Biochem Biophys Res Commun. 2018. PMID: 30448057
-
Conservation and diversification of the miR166 family in soybean and potential roles of newly identified miR166s.BMC Plant Biol. 2017 Feb 1;17(1):32. doi: 10.1186/s12870-017-0983-9. BMC Plant Biol. 2017. PMID: 28143404 Free PMC article.
-
14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean.Plant J. 2012 Jul;71(2):239-50. doi: 10.1111/j.1365-313X.2012.04986.x. Epub 2012 May 17. Plant J. 2012. PMID: 22404168
-
Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max).BMC Genomics. 2015 Nov 16;16:951. doi: 10.1186/s12864-015-2149-1. BMC Genomics. 2015. PMID: 26572792 Free PMC article.
-
Soybean proteomics for unraveling abiotic stress response mechanism.J Proteome Res. 2013 Nov 1;12(11):4670-84. doi: 10.1021/pr400604b. Epub 2013 Sep 24. J Proteome Res. 2013. PMID: 24016329 Review.
Cited by
-
Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress.Int J Mol Sci. 2018 May 31;19(6):1634. doi: 10.3390/ijms19061634. Int J Mol Sci. 2018. PMID: 29857524 Free PMC article. Review.
-
Identification and characterization of GmMYB118 responses to drought and salt stress.BMC Plant Biol. 2018 Dec 3;18(1):320. doi: 10.1186/s12870-018-1551-7. BMC Plant Biol. 2018. PMID: 30509166 Free PMC article.
-
Characterizing the Role of TaWRKY13 in Salt Tolerance.Int J Mol Sci. 2019 Nov 14;20(22):5712. doi: 10.3390/ijms20225712. Int J Mol Sci. 2019. PMID: 31739570 Free PMC article.
-
Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses.Int J Mol Sci. 2019 Nov 25;20(23):5909. doi: 10.3390/ijms20235909. Int J Mol Sci. 2019. PMID: 31775269 Free PMC article.
-
Genome-wide identification and characterization of mungbean CIRCADIAN CLOCK ASSOCIATED 1 like genes reveals an important role of VrCCA1L26 in flowering time regulation.BMC Genomics. 2022 May 17;23(1):374. doi: 10.1186/s12864-022-08620-7. BMC Genomics. 2022. PMID: 35581536 Free PMC article.
References
-
- Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G., Zhiqiang L., Yunfei Z., Xiaoxiao W., Xiaoming Q., et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

