Active Site Metal Identity Alters Histone Deacetylase 8 Substrate Selectivity: A Potential Novel Regulatory Mechanism
- PMID: 28937750
- PMCID: PMC6050578
- DOI: 10.1021/acs.biochem.7b00851
Active Site Metal Identity Alters Histone Deacetylase 8 Substrate Selectivity: A Potential Novel Regulatory Mechanism
Abstract
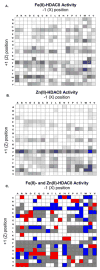
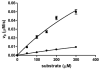
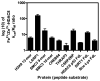
Histone deacetylase 8 (HDAC8) is a well-characterized member of the class I acetyl-lysine deacetylase (HDAC) family. Previous work has shown that the efficiency of HDAC8-catalyzed deacetylation of a methylcoumarin peptide varies depending on the identity of the divalent metal ion in the HDAC8 active site. Here we demonstrate that both HDAC8 activity and substrate selectivity for a diverse range of peptide substrates depend on the identity of the active site metal ion. Varied deacetylase activities of Fe(II)- and Zn(II)-HDAC8 toward an array of peptide substrates were identified using self-assembled monolayers for matrix-assisted laser desorption ionization (SAMDI) mass spectrometry. Subsequently, the metal dependence of deacetylation of peptides of biological interest was measured using an in vitro peptide assay. While Fe(II)-HDAC8 is generally more active than Zn(II)-HDAC8, the Fe(II)/Zn(II) HDAC8 activity ratio varies widely (from 2 to 150) among the peptides tested. These data provide support for the hypothesis that HDAC8 may undergo metal switching in vivo that, in turn, may regulate its activity. However, future studies are needed to explore the identity of the metal ion bound to HDAC8 in cells under varied conditions.
Conflict of interest statement
The authors declare no competing financial interests
Figures
Similar articles
-
Kinetics and thermodynamics of metal-binding to histone deacetylase 8.Protein Sci. 2015 Mar;24(3):354-65. doi: 10.1002/pro.2623. Epub 2015 Jan 13. Protein Sci. 2015. PMID: 25516458 Free PMC article.
-
Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion.Biochemistry. 2006 May 16;45(19):6170-8. doi: 10.1021/bi060212u. Biochemistry. 2006. PMID: 16681389
-
Histone Deacetylase 8: Characterization of Physiological Divalent Metal Catalysis.J Phys Chem B. 2016 Jul 7;120(26):5884-95. doi: 10.1021/acs.jpcb.6b00997. Epub 2016 Mar 30. J Phys Chem B. 2016. PMID: 26996235
-
Novel structural insights into class I and II histone deacetylases.Curr Top Med Chem. 2009;9(3):235-40. doi: 10.2174/156802609788085304. Curr Top Med Chem. 2009. PMID: 19355988 Review.
-
HDAC8 substrates: Histones and beyond.Biopolymers. 2013 Feb;99(2):112-26. doi: 10.1002/bip.22135. Biopolymers. 2013. PMID: 23175386 Free PMC article. Review.
Cited by
-
Repressing iron overload ameliorates central post-stroke pain via the Hdac2-Kv1.2 axis in a rat model of hemorrhagic stroke.Neural Regen Res. 2024 Dec 1;19(12):2708-2722. doi: 10.4103/NRR.NRR-D-23-01498. Epub 2024 Apr 1. Neural Regen Res. 2024. PMID: 38595289 Free PMC article.
-
The enzyme activity of histone deacetylase 8 is modulated by a redox-switch.Redox Biol. 2019 Jan;20:60-67. doi: 10.1016/j.redox.2018.09.013. Epub 2018 Sep 27. Redox Biol. 2019. PMID: 30292946 Free PMC article.
-
Continuous Activity Assay for HDAC11 Enabling Reevaluation of HDAC Inhibitors.ACS Omega. 2019 Nov 15;4(22):19895-19904. doi: 10.1021/acsomega.9b02808. eCollection 2019 Nov 26. ACS Omega. 2019. PMID: 31788622 Free PMC article.
-
A distal regulatory region of a class I human histone deacetylase.Nat Commun. 2020 Jul 31;11(1):3841. doi: 10.1038/s41467-020-17610-w. Nat Commun. 2020. PMID: 32737323 Free PMC article.
-
Phosphorylation of Histone Deacetylase 8: Structural and Mechanistic Analysis of the Phosphomimetic S39E Mutant.Biochemistry. 2019 Nov 12;58(45):4480-4493. doi: 10.1021/acs.biochem.9b00653. Epub 2019 Nov 4. Biochemistry. 2019. PMID: 31633931 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

