Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis
- PMID: 28937985
- PMCID: PMC5627961
- DOI: 10.1371/journal.pgen.1007022
Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis
Abstract
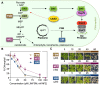
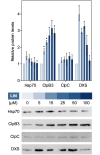
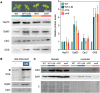
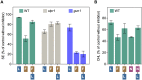
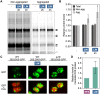
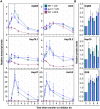
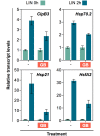
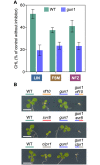
Disruption of protein homeostasis in chloroplasts impairs the correct functioning of essential metabolic pathways, including the methylerythritol 4-phosphate (MEP) pathway for the production of plastidial isoprenoids involved in photosynthesis and growth. We previously found that misfolded and aggregated forms of the first enzyme of the MEP pathway are degraded by the Clp protease with the involvement of Hsp70 and Hsp100/ClpC1 chaperones in Arabidopsis thaliana. By contrast, the combined unfolding and disaggregating actions of Hsp70 and Hsp100/ClpB3 chaperones allow solubilization and hence reactivation of the enzyme. The repair pathway is promoted when the levels of ClpB3 proteins increase upon reduction of Clp protease activity in mutants or wild-type plants treated with the chloroplast protein synthesis inhibitor lincomycin (LIN). Here we show that LIN treatment rapidly increases the levels of aggregated proteins in the chloroplast, unleashing a specific retrograde signaling pathway that up-regulates expression of ClpB3 and other nuclear genes encoding plastidial chaperones. As a consequence, folding capacity is increased to restore protein homeostasis. This sort of chloroplast unfolded protein response (cpUPR) mechanism appears to be mediated by the heat shock transcription factor HsfA2. Expression of HsfA2 and cpUPR-related target genes is independent of GUN1, a central integrator of retrograde signaling pathways. However, double mutants defective in both GUN1 and plastome gene expression (or Clp protease activity) are seedling lethal, confirming that the GUN1 protein is essential for protein homeostasis in chloroplasts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Grimm B, Dehesh K, Zhang L, Leister D (2014) Intracellular communication. Mol Plant 7: 1071–1074. doi: 10.1093/mp/ssu073 - DOI - PubMed
-
- Kleine T, Leister D (2016) Retrograde signaling: Organelles go networking. Biochim Biophys Acta 1857: 1313–1325. doi: 10.1016/j.bbabio.2016.03.017 - DOI - PubMed
-
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annu Rev Plant Biol 67: 25–53. doi: 10.1146/annurev-arplant-043015-111854 - DOI - PubMed
-
- Sun AZ, Guo FQ (2016) Chloroplast Retrograde Regulation of Heat Stress Responses in Plants. Front Plant Sci 7: 398 doi: 10.3389/fpls.2016.00398 - DOI - PMC - PubMed
-
- Lin YF, Haynes CM (2016) Metabolism and the UPR(mt). Mol Cell 61: 677–682. doi: 10.1016/j.molcel.2016.02.004 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

