Emerging Opportunities for Target Discovery in Rare Cancers
- PMID: 28938087
- PMCID: PMC5857178
- DOI: 10.1016/j.chembiol.2017.08.002
Emerging Opportunities for Target Discovery in Rare Cancers
Abstract
Rare cancers pose unique challenges to research due to their low incidence. Barriers include a scarcity of tissue and experimental models to enable basic research and insufficient patient accrual for clinical studies. Consequently, an understanding of the genetic and cellular features of many rare cancer types and their associated vulnerabilities has been lacking. However, new opportunities are emerging to facilitate discovery of therapeutic targets in rare cancers. Online platforms are allowing patients with rare cancers to organize on an unprecedented scale, tumor genome sequencing is now routinely performed in research and clinical settings, and the efficiency of patient-derived model generation has improved. New CRISPR/Cas9 and small-molecule libraries permit cancer dependency discovery in a rapid and systematic fashion. In parallel, large-scale studies of common cancers now provide reference datasets to help interpret rare cancer profiling data. Together, these advances motivate consideration of new research frameworks to accelerate rare cancer target discovery.
Keywords: CRISPR/Cas9; chordoma; conditionally reprogrammed cells; genomics; next-generation sequencing; organoid; patient-derived xenograft; rare cancer; small-molecule screen; target discovery.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
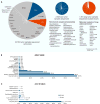


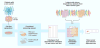

References
-
- Abrams J, Conley B, Mooney M, Zwiebel J, Chen A, Welch JJ, Takebe N, Malik S, McShane L, Korn E, et al. National Cancer Institute’s Precision Medicine Initiatives for the new National Clinical Trials Network. Am Soc Clin Oncol Educ Book. 2014:71–76. - PubMed
-
- Afani L, Errihani H, Benchafai I, Lalami Y. Nasopharyngeal adenoid cystic carcinoma, a rare but highly challenging disease with unmet therapeutic needs: a case-report and review of the literature. Cancer Radiother. 2016;20:400–404. - PubMed
-
- American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–448. - PubMed
-
- Arıkan M, Togral G, Hastürk AE, Aktaş E, Güngör S. Management and retrospective analysis of primary and metastatic sacral tumors and infections: evaluation with 73 cases. Eklem Hastalik Cerrahisi. 2014;25:126–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

