Small-Molecule Inhibitors Targeting DNA Repair and DNA Repair Deficiency in Research and Cancer Therapy
- PMID: 28938088
- PMCID: PMC5679738
- DOI: 10.1016/j.chembiol.2017.08.027
Small-Molecule Inhibitors Targeting DNA Repair and DNA Repair Deficiency in Research and Cancer Therapy
Abstract
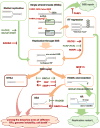
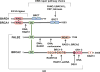
To maintain stable genomes and to avoid cancer and aging, cells need to repair a multitude of deleterious DNA lesions, which arise constantly in every cell. Processes that support genome integrity in normal cells, however, allow cancer cells to develop resistance to radiation and DNA-damaging chemotherapeutics. Chemical inhibition of the key DNA repair proteins and pharmacologically induced synthetic lethality have become instrumental in both dissecting the complex DNA repair networks and as promising anticancer agents. The difficulty in capitalizing on synthetically lethal interactions in cancer cells is that many potential targets do not possess well-defined small-molecule binding determinates. In this review, we discuss several successful campaigns to identify and leverage small-molecule inhibitors of the DNA repair proteins, from PARP1, a paradigm case for clinically successful small-molecule inhibitors, to coveted new targets, such as RAD51 recombinase, RAD52 DNA repair protein, MRE11 nuclease, and WRN DNA helicase.
Keywords: BLM DNA helicase; BRCA1; BRCA2; DNA repair; MRE11; PARP inhibitors; PARP1 (poly(ADP-ribose)polymerase1); RAD51; RAD52; RECQ1 DNA helicase; TOP1; WRN DNA helicase; homologous recombination.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures


References
-
- Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1525–1530. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

