Pioneer Factors FOXA1 and FOXA2 Assist Selective Glucocorticoid Receptor Signaling in Human Endometrial Cells
- PMID: 28938408
- PMCID: PMC5695839
- DOI: 10.1210/en.2017-00361
Pioneer Factors FOXA1 and FOXA2 Assist Selective Glucocorticoid Receptor Signaling in Human Endometrial Cells
Abstract
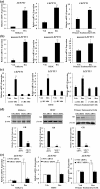
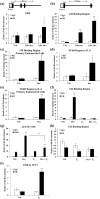
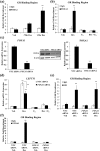
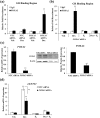
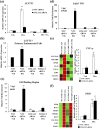
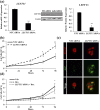
Successful pregnancy relies on dynamic control of cell signaling to achieve uterine receptivity and the necessary biological changes required for endometrial decidualization, embryo implantation, and fetal development. Glucocorticoids are master regulators of intracellular signaling and can directly regulate embryo implantation and endometrial remodeling during murine pregnancy. In immortalized human uterine cells, we have shown that glucocorticoids and estradiol (E2) coregulate thousands of genes. Recently, glucocorticoids and E2 were shown to coregulate the expression of Left-right determination factor 1 (LEFTY1), previously implicated in the regulation of decidualization. To elucidate the molecular mechanism by which glucocorticoids and E2 regulate the expression of LEFTY1, immortalized and primary human endometrial cells were evaluated for gene expression and receptor recruitment to regulatory regions of the LEFTY1 gene. Glucocorticoid administration induced expression of LEFTY1 messenger RNA and protein and recruitment of the glucocorticoid receptor (GR) and activated polymerase 2 to the promoter of LEFTY1. Glucocorticoid-mediated recruitment of GR was dependent on pioneer factors FOXA1 and FOXA2. E2 was found to antagonize glucocorticoid-mediated induction of LEFTY1 by reducing recruitment of GR, FOXA1, FOXA2, and activated polymerase 2 to the LEFTY1 promoter. Gene expression analysis identified several genes whose glucocorticoid-dependent induction required FOXA1 and FOXA2 in endometrial cells. These results suggest a molecular mechanism by which E2 antagonizes GR-dependent induction of specific genes by preventing the recruitment of the pioneer factors FOXA1 and FOXA2 in a physiologically relevant model.
Copyright © 2017 Endocrine Society.
Figures


Similar articles
-
Interaction of glucocorticoid receptor (GR) with estrogen receptor (ER) α and activator protein 1 (AP1) in dexamethasone-mediated interference of ERα activity.J Biol Chem. 2013 Aug 16;288(33):24020-34. doi: 10.1074/jbc.M113.473819. Epub 2013 Jun 28. J Biol Chem. 2013. PMID: 23814048 Free PMC article.
-
Cell-specific expression of ENACα gene by FOXA1 in the glucocorticoid receptor pathway.Int J Immunopathol Pharmacol. 2020 Jan-Dec;34:2058738420946192. doi: 10.1177/2058738420946192. Int J Immunopathol Pharmacol. 2020. PMID: 32838581 Free PMC article.
-
Foxa1 and Foxa2 regulate α-cell differentiation, glucagon biosynthesis, and secretion.Endocrinology. 2014 Oct;155(10):3781-92. doi: 10.1210/en.2013-1843. Epub 2014 Jul 24. Endocrinology. 2014. PMID: 25057789
-
Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance.Endocr Dev. 2013;24:41-56. doi: 10.1159/000342502. Epub 2013 Feb 1. Endocr Dev. 2013. PMID: 23392094 Free PMC article. Review.
-
Interplay of estrogen receptors and FOXA factors in the liver cancer.Mol Cell Endocrinol. 2015 Dec 15;418 Pt 3(0 3):334-9. doi: 10.1016/j.mce.2015.01.043. Epub 2015 Feb 4. Mol Cell Endocrinol. 2015. PMID: 25661537 Free PMC article. Review.
Cited by
-
Tissue-specific progesterone receptor-chromatin binding and the regulation of progesterone-dependent gene expression.Sci Rep. 2019 Aug 19;9(1):11966. doi: 10.1038/s41598-019-48333-8. Sci Rep. 2019. PMID: 31427604 Free PMC article.
-
Integrative analysis of the forkhead box A2 (FOXA2) cistrome for the human endometrium.FASEB J. 2019 Jul;33(7):8543-8554. doi: 10.1096/fj.201900013R. Epub 2019 Apr 5. FASEB J. 2019. PMID: 30951376 Free PMC article.
-
Seminal Plasma Triggers the Differential Expression of the Glucocorticoid Receptor (NR3C1/GR) in the Rabbit Reproductive Tract.Animals (Basel). 2020 Nov 19;10(11):2158. doi: 10.3390/ani10112158. Animals (Basel). 2020. PMID: 33228207 Free PMC article.
-
Stress, Sex, and Sugar: Glucocorticoids and Sex-Steroid Crosstalk in the Sex-Specific Misprogramming of Metabolism.J Endocr Soc. 2020 Jul 3;4(8):bvaa087. doi: 10.1210/jendso/bvaa087. eCollection 2020 Aug 1. J Endocr Soc. 2020. PMID: 32734132 Free PMC article. Review.
-
Homeobox regulator Wilms Tumour 1 is displaced by androgen receptor at cis-regulatory elements in the endometrium of PCOS patients.Front Endocrinol (Lausanne). 2024 Apr 30;15:1368494. doi: 10.3389/fendo.2024.1368494. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38745948 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

