Transcriptomic profiles of human foreskin fibroblast cells in response to orf virus
- PMID: 28938587
- PMCID: PMC5601683
- DOI: 10.18632/oncotarget.17417
Transcriptomic profiles of human foreskin fibroblast cells in response to orf virus
Abstract
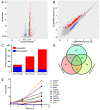
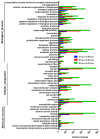
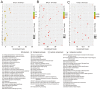
Orf virus has been utilized as a safe and efficient viral vector against not only diverse infectious diseases, but also against tumors. However, the nature of the genes triggered by the vector in human cells is poorly characterized. Using RNA sequencing technology, we compared specific changes in the transcriptomic profiles in human foreskin fibroblast cells following infection by the orf virus. The results indicated that orf virus upregulates or downregulates expression of a variety of genes, including genes involved in antiviral immune response, apoptosis, cell cycle and a series of signaling pathways, such as the IFN and p53-signaling pathways. The orf virus stimulates or inhibits immune gene expression such as chemokines, chemokine receptors, cytokines, cytokine receptors, and molecules involved in antigen uptake and processing after infection. Expression of pro-apoptotic genes increased at 8 hours post-infection. The p53 signaling pathway was activated to induce apoptosis at the same time. However, the cell cycle program was promoted after infection, which may be due to the immunomodulatory genes of the orf virus. This presents the first description of transcription profile changes in human foreskin fibroblast cells after orf virus infection and provides an in-depth analysis of the interaction between the host and orf virus. These data offer new insights into the understanding of the mechanisms of infection by orf virus and identify potential targets for future studies.
Keywords: antiviral immune response; apoptosis; cell cycle; orf virus; transcriptomic profiles.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures





Similar articles
-
Transcriptional analysis of immune-related gene expression in p53-deficient mice with increased susceptibility to influenza A virus infection.BMC Med Genomics. 2015 Aug 18;8:52. doi: 10.1186/s12920-015-0127-8. BMC Med Genomics. 2015. PMID: 26282854 Free PMC article.
-
ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection.J Virol. 2005 Aug;79(16):10308-29. doi: 10.1128/JVI.79.16.10308-10329.2005. J Virol. 2005. PMID: 16051824 Free PMC article.
-
Orf virus infection and host immunity.Curr Opin Infect Dis. 2006 Apr;19(2):127-31. doi: 10.1097/01.qco.0000216622.75326.ef. Curr Opin Infect Dis. 2006. PMID: 16514336 Review.
-
Prevention of virus persistence and protection against immunopathology after Borna disease virus infection of the brain by a novel Orf virus recombinant.J Virol. 2005 Jan;79(1):314-25. doi: 10.1128/JVI.79.1.314-325.2005. J Virol. 2005. PMID: 15596826 Free PMC article.
-
Subversion and piracy: DNA viruses and immune evasion.Res Vet Sci. 2001 Jun;70(3):205-19. doi: 10.1053/rvsc.2001.0462. Res Vet Sci. 2001. PMID: 11676616 Review.
Cited by
-
Antibody-dependent enhancement of ORFV uptake into host cells.Virulence. 2025 Dec;16(1):2466503. doi: 10.1080/21505594.2025.2466503. Epub 2025 Feb 15. Virulence. 2025. PMID: 39954287 Free PMC article.
-
Integrative Transcriptomic Network Analysis of Butyrate Treated Colorectal Cancer Cells.Cancers (Basel). 2021 Feb 5;13(4):636. doi: 10.3390/cancers13040636. Cancers (Basel). 2021. PMID: 33562636 Free PMC article.
-
Evolution-Driven Attenuation of Alphaviruses Highlights Key Glycoprotein Determinants Regulating Viral Infectivity and Dissemination.Cell Rep. 2019 Jul 9;28(2):460-471.e5. doi: 10.1016/j.celrep.2019.06.022. Cell Rep. 2019. PMID: 31291581 Free PMC article.
-
Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis.Biochem J. 2020 Dec 11;477(23):4527-4541. doi: 10.1042/BCJ20200776. Biochem J. 2020. PMID: 33175095 Free PMC article.
-
The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: possible implication as theranostic agents.RNA Biol. 2021 Jan;18(1):1-15. doi: 10.1080/15476286.2020.1790198. Epub 2020 Jul 13. RNA Biol. 2021. PMID: 32615049 Free PMC article. Review.
References
-
- Spyrou V, Valiakos G. Orf virus infection in sheep or goats. Vet Microbiol. 2015;181:178–82. - PubMed
-
- Büttner M, Rziha HJ. Parapoxviruses: from the lesion to the viral genome. J Vet Med B Infect Dis Vet Public Health. 2002;49:7–16. - PubMed
-
- Thurman RJ, Fitch RW. Images in clinical medicine. Contagious ecthyma. N Engl J Med. 2015;372:e12. - PubMed
-
- Rajkomar V, Hannah M, Coulson IH, Owen CM. A case of human to human transmission of orf between mother and child. Clin Exp Dermatol. 2016;41:60–63. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

