Potential independent action of sigma receptor ligands through inhibition of the Kv2.1 channel
- PMID: 28938641
- PMCID: PMC5601737
- DOI: 10.18632/oncotarget.19581
Potential independent action of sigma receptor ligands through inhibition of the Kv2.1 channel
Abstract
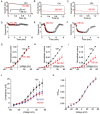
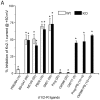
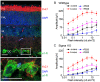
The sigma-1 receptor (σ1-R) and sigma-2 receptor (σ2-R) are potential drug targets for treatment of cancer, pain, depression, retinal degeneration and other neuronal diseases. Previous reports show that sigma-1 receptor modulates the activities of multiple channels. We are interested in possible sigma receptor modulation of Kv2.1, a K+ channel abundant in retinal photoreceptors. We tested the effect of established sigma receptor ligands on Kv2.1 channels which were stably expressed in HEK293 cells. Surprisingly, σ1-R antagonists inhibited Kv2.1 currents in both wild type and σ1-R knockout HEK293 cells that we engineered using the CRISPR/Cas9 technology. Moreover, PB28, a σ1-R antagonist and also σ2-R agonist, inhibited Kv2.1 in σ1-R knockout cells, but this action was not blocked by the σ2-R antagonists that did not have an effect on Kv2.1. We also observed inhibition of electroretinogram by PB28 in wild type as well as σ1-R knockout mice. Thus, the results in this study indicate that the Kv2.1-inhibiting function of the sigma ligands is not sigma receptor dependent, suggesting a direct effect of these ligands on the Kv2.1 channel.
Keywords: CRISPR/Cas9; Kv2.1 channel; electroretinogram; patch-clamp; sigma receptor ligands.
Conflict of interest statement
CONFLICTS OF INTEREST None.
Figures
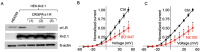


Similar articles
-
Antiproliferative and cytotoxic effects of some sigma2 agonists and sigma1 antagonists in tumour cell lines.Naunyn Schmiedebergs Arch Pharmacol. 2004 Aug;370(2):106-13. doi: 10.1007/s00210-004-0961-2. Epub 2004 Jul 31. Naunyn Schmiedebergs Arch Pharmacol. 2004. PMID: 15322732
-
Cyclohexylpiperazine derivative PB28, a sigma2 agonist and sigma1 antagonist receptor, inhibits cell growth, modulates P-glycoprotein, and synergizes with anthracyclines in breast cancer.Mol Cancer Ther. 2006 Jul;5(7):1807-16. doi: 10.1158/1535-7163.MCT-05-0402. Mol Cancer Ther. 2006. PMID: 16891467
-
The attenuation of learning impairments induced after exposure to CO or trimethyltin in mice by sigma (sigma) receptor ligands involves both sigma1 and sigma2 sites.Br J Pharmacol. 1999 May;127(2):335-42. doi: 10.1038/sj.bjp.0702553. Br J Pharmacol. 1999. PMID: 10385231 Free PMC article.
-
Introduction to Sigma Proteins: Evolution of the Concept of Sigma Receptors.Handb Exp Pharmacol. 2017;244:1-11. doi: 10.1007/164_2017_41. Handb Exp Pharmacol. 2017. PMID: 28871306 Review.
-
Small-Molecule Modulators of Sigma1 and Sigma2/TMEM97 in the Context of Cancer: Foundational Concepts and Emerging Themes.Front Pharmacol. 2019 Oct 21;10:1141. doi: 10.3389/fphar.2019.01141. eCollection 2019. Front Pharmacol. 2019. PMID: 31695608 Free PMC article. Review.
Cited by
-
Role of Sigma Receptors in Alcohol Addiction.Front Pharmacol. 2019 Jun 14;10:687. doi: 10.3389/fphar.2019.00687. eCollection 2019. Front Pharmacol. 2019. PMID: 31258483 Free PMC article. Review.
-
The sigma-1 receptor as a regulator of dopamine neurotransmission: A potential therapeutic target for methamphetamine addiction.Pharmacol Ther. 2018 Jun;186:152-167. doi: 10.1016/j.pharmthera.2018.01.009. Epub 2018 Jan 31. Pharmacol Ther. 2018. PMID: 29360540 Free PMC article. Review.
-
Sigmar1's Molecular, Cellular, and Biological Functions in Regulating Cellular Pathophysiology.Front Physiol. 2021 Jul 7;12:705575. doi: 10.3389/fphys.2021.705575. eCollection 2021. Front Physiol. 2021. PMID: 34305655 Free PMC article. Review.
-
Retinoschisin and novel Na/K-ATPase interaction partners Kv2.1 and Kv8.2 define a growing protein complex at the inner segments of mammalian photoreceptors.Cell Mol Life Sci. 2022 Jul 25;79(8):448. doi: 10.1007/s00018-022-04409-9. Cell Mol Life Sci. 2022. PMID: 35876901 Free PMC article.
-
Is the sigma-1 receptor a potential pharmacological target for cardiac pathologies? A systematic review.Int J Cardiol Heart Vasc. 2019 Dec 28;26:100449. doi: 10.1016/j.ijcha.2019.100449. eCollection 2020 Feb. Int J Cardiol Heart Vasc. 2019. PMID: 31909177 Free PMC article. Review.
References
-
- Quirion R, Bowen WD, Itzhak Y, Junien JL, Musacchio JM, Rothman RB, Su TP, Tam SW, Taylor DP. A proposal for the classification of sigma binding sites. Trends Pharmacol Sci. 1992;13:85–6. - PubMed
-
- Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. https://doi.org/10.16/j.cell.2007.08.036. - DOI - PubMed
-
- Chu UB, Mavlyutov TA, Chu ML, Yang H, Schulman A, Mesangeau C, McCurdy CR, Guo LW, Ruoho AE. The sigma-2 receptor and progesterone receptor membrane component 1 are different binding sites derived from independent genes. EBioMedicine. 2015;2:1806–13. https://doi.org/10.1016/j.ebiom.2015.10.017. - DOI - PMC - PubMed
-
- Su TP, Hayashi T, Maurice T, Buch S, Ruoho AE. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol Sci. 2010;31:557–66. https://doi.org/10.16/j.tips.2010.08.007. - DOI - PMC - PubMed
-
- Nguyen L, Lucke-Wold BP, Mookerjee SA, Cavendish JZ, Robson MJ, Scandinaro AL, Matsumoto RR. Role of sigma-1 receptors in neurodegenerative diseases. J Pharmacol Sci. 2015;127:17–29. https://doi.org/10.1016/j.jphs.2014.12.005. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

