Metagenomic characterization of ambulances across the USA
- PMID: 28938903
- PMCID: PMC5610413
- DOI: 10.1186/s40168-017-0339-6
Metagenomic characterization of ambulances across the USA
Abstract
Background: Microbial communities in our built environments have great influence on human health and disease. A variety of built environments have been characterized using a metagenomics-based approach, including some healthcare settings. However, there has been no study to date that has used this approach in pre-hospital settings, such as ambulances, an important first point-of-contact between patients and hospitals.
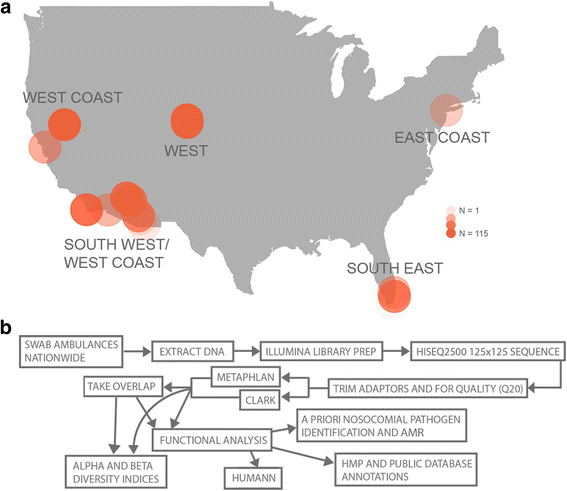
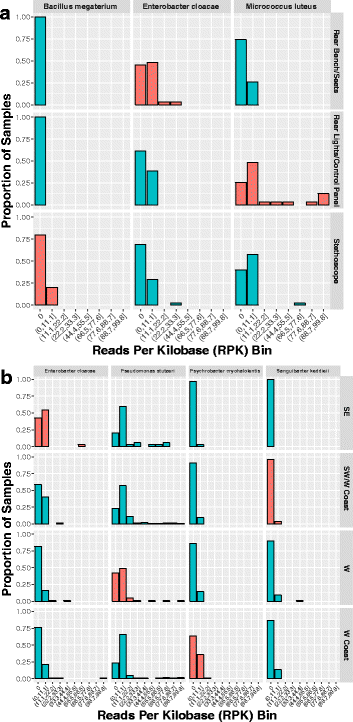
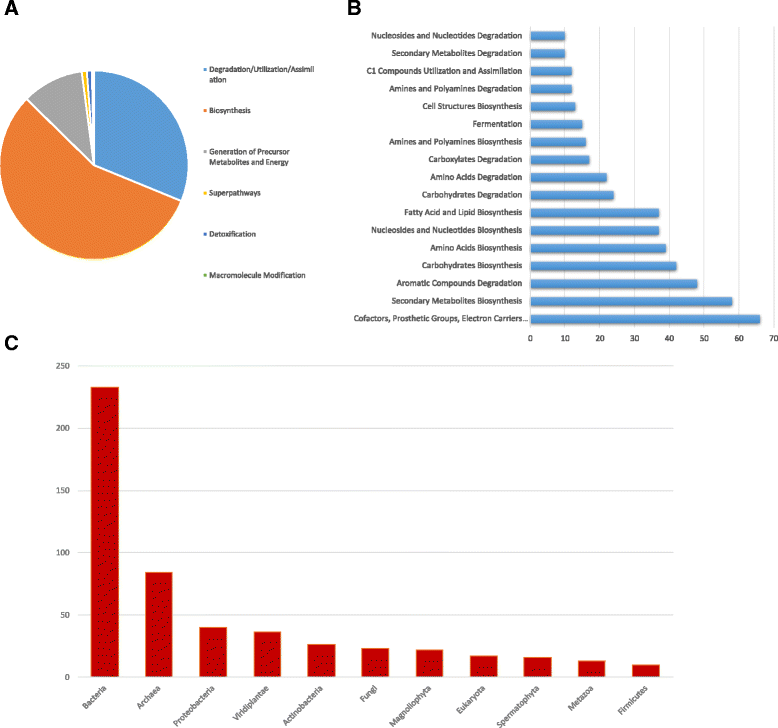
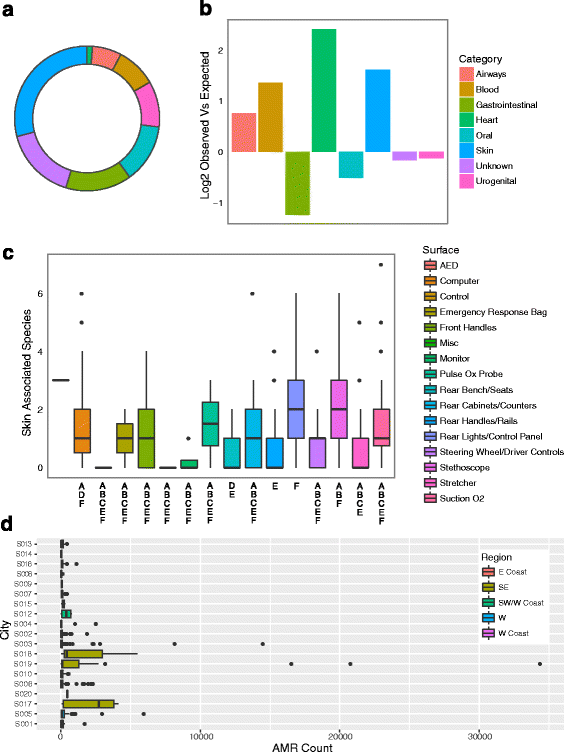
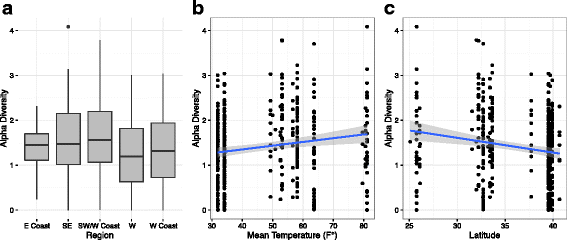
Results: We sequenced 398 samples from 137 ambulances across the USA using shotgun sequencing. We analyzed these data to explore the microbial ecology of ambulances including characterizing microbial community composition, nosocomial pathogens, patterns of diversity, presence of functional pathways and antimicrobial resistance, and potential spatial and environmental factors that may contribute to community composition. We found that the top 10 most abundant species are either common built environment microbes, microbes associated with the human microbiome (e.g., skin), or are species associated with nosocomial infections. We also found widespread evidence of antimicrobial resistance markers (hits ~ 90% samples). We identified six factors that may influence the microbial ecology of ambulances including ambulance surfaces, geographical-related factors (including region, longitude, and latitude), and weather-related factors (including temperature and precipitation).
Conclusions: While the vast majority of microbial species classified were beneficial, we also found widespread evidence of species associated with nosocomial infections and antimicrobial resistance markers. This study indicates that metagenomics may be useful to characterize the microbial ecology of pre-hospital ambulance settings and that more rigorous testing and cleaning of ambulances may be warranted.
Keywords: Ambulance; Antimicrobial resistance; Classification; Hospital-acquired infections; Metagenomics; Microbial ecology; Nosocomial pathogens; Pre-hospital setting; Taxonomy; Whole-genome sequencing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable for new data collected. HMP data was acquired from their public database.
Consent for publication
Not applicable
Competing interests
NO, RO, and CM hold shares in a company that builds technology to survey hospital environments to identify pathogens, however that company’s technology is not used in this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Hospital Microbiome Variations As Analyzed by High-Throughput Sequencing.OMICS. 2019 Sep;23(9):426-438. doi: 10.1089/omi.2019.0111. Epub 2019 Aug 8. OMICS. 2019. PMID: 31393213
-
Whole metagenome profiles of particulates collected from the International Space Station.Microbiome. 2017 Jul 17;5(1):81. doi: 10.1186/s40168-017-0292-4. Microbiome. 2017. PMID: 28716113 Free PMC article.
-
Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details.Microbiome. 2017 Feb 10;5(1):20. doi: 10.1186/s40168-017-0234-1. Microbiome. 2017. PMID: 28187782 Free PMC article.
-
Hospital-associated microbiota and implications for nosocomial infections.Trends Mol Med. 2015 Jul;21(7):427-32. doi: 10.1016/j.molmed.2015.03.005. Epub 2015 Apr 20. Trends Mol Med. 2015. PMID: 25907678 Review.
-
Practical considerations for sampling and data analysis in contemporary metagenomics-based environmental studies.J Microbiol Methods. 2018 Nov;154:14-18. doi: 10.1016/j.mimet.2018.09.020. Epub 2018 Oct 1. J Microbiol Methods. 2018. PMID: 30287354 Review.
Cited by
-
Targeted Hybridization Capture of SARS-CoV-2 and Metagenomics Enables Genetic Variant Discovery and Nasal Microbiome Insights.Microbiol Spectr. 2021 Oct 31;9(2):e0019721. doi: 10.1128/Spectrum.00197-21. Epub 2021 Sep 1. Microbiol Spectr. 2021. PMID: 34468193 Free PMC article.
-
Rapid metagenomics analysis of EMS vehicles for monitoring pathogen load using nanopore DNA sequencing.PLoS One. 2019 Jul 24;14(7):e0219961. doi: 10.1371/journal.pone.0219961. eCollection 2019. PLoS One. 2019. PMID: 31339905 Free PMC article.
-
An improved workflow for accurate and robust healthcare environmental surveillance using metagenomics.Microbiome. 2022 Dec 2;10(1):206. doi: 10.1186/s40168-022-01412-x. Microbiome. 2022. PMID: 36457108 Free PMC article.
-
Ozone Disinfection for Elimination of Bacteria and Degradation of SARS-CoV2 RNA for Medical Environments.Genes (Basel). 2022 Dec 28;14(1):85. doi: 10.3390/genes14010085. Genes (Basel). 2022. PMID: 36672826 Free PMC article.
-
Comparative genomics of Bacteria commonly identified in the built environment.BMC Genomics. 2019 Jan 28;20(1):92. doi: 10.1186/s12864-018-5389-z. BMC Genomics. 2019. PMID: 30691394 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

