The F-Box Protein ZYGO1 Mediates Bouquet Formation to Promote Homologous Pairing, Synapsis, and Recombination in Rice Meiosis
- PMID: 28939596
- PMCID: PMC5774573
- DOI: 10.1105/tpc.17.00287
The F-Box Protein ZYGO1 Mediates Bouquet Formation to Promote Homologous Pairing, Synapsis, and Recombination in Rice Meiosis
Abstract
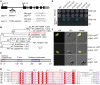
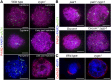
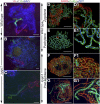
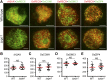
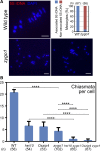
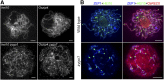
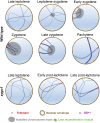
Telomere bouquet formation, a highly conserved meiotic event, plays an important role in homologous pairing and therefore progression of meiosis; however, the underlying molecular mechanism remains largely unknown. Here, we identified ZYGOTENE1 (ZYGO1), a novel F-box protein in rice (Oryza sativa), and verified its essential role in bouquet formation during early meiosis. In zygo1 mutants, zygotene chromosome aggregation and telomere clustering failed to occur. The suppressed telomere clustering in homologous pairing aberration in rice meiosis1 (pair1) zygo1 and rice completion of meiotic recombination (Oscom1) zygo1 double mutants, together with the altered localization of OsSAD1 (a SUN protein associated with the nuclear envelope) in zygo1, showed that ZYGO1 has a significant function in bouquet formation. In addition, the interaction between ZYGO1 and rice SKP1-like protein 1 suggested that ZYGO1 might modulate bouquet formation as a component of the SKP1-Cullin1-F-box complex. Although double-strand break formation and early recombination element installation occurred normally, zygo1 mutants showed defects in full-length pairing and synaptonemal complex assembly. Furthermore, crossover (CO) formation was disturbed, and foci of Human enhancer of invasion 10 were restricted to the partially synapsed chromosome regions, indicating that CO reduction might be caused by the failure of full-length chromosome alignment in zygo1 Therefore, we propose that ZYGO1 mediates bouquet formation to efficiently promote homolog pairing, synapsis, and CO formation in rice meiosis.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures

Similar articles
-
OsHOP2 regulates the maturation of crossovers by promoting homologous pairing and synapsis in rice meiosis.New Phytol. 2019 Apr;222(2):805-819. doi: 10.1111/nph.15664. Epub 2019 Jan 25. New Phytol. 2019. PMID: 30584664
-
The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis.Plant Cell. 2004 Apr;16(4):1008-20. doi: 10.1105/tpc.020701. Epub 2004 Mar 18. Plant Cell. 2004. PMID: 15031413 Free PMC article.
-
The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis.Plant J. 2012 Oct;72(1):18-30. doi: 10.1111/j.1365-313X.2012.05025.x. Epub 2012 Aug 1. Plant J. 2012. PMID: 22507309
-
A bouquet of chromosomes.J Cell Sci. 2004 Aug 15;117(Pt 18):4025-32. doi: 10.1242/jcs.01363. J Cell Sci. 2004. PMID: 15316078 Review.
-
The cytogenetics of homologous chromosome pairing in meiosis in plants.Cytogenet Genome Res. 2008;120(3-4):313-9. doi: 10.1159/000121080. Epub 2008 May 23. Cytogenet Genome Res. 2008. PMID: 18504360 Review.
Cited by
-
FBXO47 regulates telomere-inner nuclear envelope integration by stabilizing TRF2 during meiosis.Nucleic Acids Res. 2019 Dec 16;47(22):11755-11770. doi: 10.1093/nar/gkz992. Nucleic Acids Res. 2019. PMID: 31724724 Free PMC article.
-
The SUN Domain Proteins OsSUN1 and OsSUN2 Play Critical but Partially Redundant Roles in Meiosis.Plant Physiol. 2020 Aug;183(4):1517-1530. doi: 10.1104/pp.20.00140. Epub 2020 Jun 18. Plant Physiol. 2020. PMID: 32554471 Free PMC article.
-
SCFRMF mediates degradation of the meiosis-specific recombinase DMC1.Nat Commun. 2023 Aug 19;14(1):5044. doi: 10.1038/s41467-023-40799-5. Nat Commun. 2023. PMID: 37598222 Free PMC article.
-
Advancing knowledge of the plant nuclear periphery and its application for crop science.Nucleus. 2020 Dec;11(1):347-363. doi: 10.1080/19491034.2020.1838697. Nucleus. 2020. PMID: 33295233 Free PMC article. Review.
-
Telomeres and Subtelomeres Dynamics in the Context of Early Chromosome Interactions During Meiosis and Their Implications in Plant Breeding.Front Plant Sci. 2021 Jun 4;12:672489. doi: 10.3389/fpls.2021.672489. eCollection 2021. Front Plant Sci. 2021. PMID: 34149773 Free PMC article. Review.
References
-
- Cheng Z. (2013). Analyzing meiotic chromosomes in rice. Methods Mol. Biol. 990: 125–134. - PubMed
-
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. (2006). Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

