Alterations in gp37 Expand the Host Range of a T4-Like Phage
- PMID: 28939606
- PMCID: PMC5691408
- DOI: 10.1128/AEM.01576-17
Alterations in gp37 Expand the Host Range of a T4-Like Phage
Abstract
The use of phages as antibacterial agents is limited by their generally narrow host ranges. The aim of this study was to make a T4-like phage, WG01, obtain the host range of another T4-like phage, QL01, by replacing its host-determinant gene region with that of QL01. This process triggered a direct expansion of the WG01 host range. The offspring of WG01 obtained the host ranges of both QL01 and WG01, as well as the ability to infect eight additional host bacteria in comparison to the wild-type strains. WQD had the widest host range; therefore, the corresponding fragments, named QD, could be used for constructing a homologous sequence library. Moreover, after a sequencing analysis of gene 37, we identified two different mechanisms responsible for the expanded host range: (i) the first generation of WG01 formed chimeras without mutations, and (ii) the second generation of WG01 mutants formed from the chimeras. The expansion of the host range indicated that regions other than the C-terminal region may indirectly change the receptor specificity by altering the supportive capacity of the binding site. Additionally, we also found the novel means by which subsequent generations expanded their host ranges, namely, by exchanging gene 37 to acquire a wider temperature range for lysis. The method developed in this work offers a quick way to change or expand the host range of a phage. Future clinical applications for screening phages against a given clinical isolate could be achieved after acquiring more suitable homologous sequences.IMPORTANCE T4-like phages have been established as safe in numerous phage therapy applications. The primary drawbacks to the use of phages as therapeutic agents include their highly specific host ranges. Thus, changing or expanding the host range of T4-like phages is beneficial for selecting phages for phage therapy. In this study, the host range of the T4-like phage WG01 was expanded using genetic manipulation. The WG01 derivatives acquired a novel means of expanding their host ranges by acquiring a wider temperature range for lysis. A region was located that had the potential to be used as a sequence region for homologous sequence recombination.
Keywords: Escherichia coli; T4-like phages; gp37; homologous recombination; host range expansion.
Copyright © 2017 American Society for Microbiology.
Figures
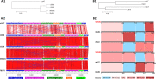




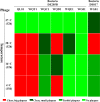
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

