Regulation of mitotic spindle assembly factor NuMA by Importin-β
- PMID: 28939615
- PMCID: PMC5674899
- DOI: 10.1083/jcb.201705168
Regulation of mitotic spindle assembly factor NuMA by Importin-β
Abstract
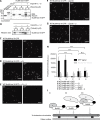
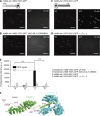
Ran-guanosine triphosphatase orchestrates mitotic spindle assembly by modulation of the interaction between Importin-α/-β and spindle assembly factors (SAFs). The inhibition of SAFs performed by importins needs to be done without much sequestration from abundant nuclear localization signal (NLS) -containing proteins. However, the molecular mechanisms that determine NLS-binding selectivity and that inhibit activity of Importin-β-regulated SAFs (e.g., nuclear mitotic apparatus protein [NuMA]) remain undefined. Here, we present a crystal structure of the Importin-α-NuMA C terminus complex showing a novel binding pattern that accounts for selective NLS recognition. We demonstrate that, in the presence of Importin-α, Importin-β inhibits the microtubule-binding function of NuMA. Further, we have identified a high-affinity microtubule-binding region that lies carboxyl-terminal to the NLS, which is sterically masked by Importin-β on being bound by Importin-α. Our study provides mechanistic evidence of how Importin-α/-β regulates the NuMA functioning required for assembly of higher-order microtubule structures, further illuminating how Ran-governed transport factors regulate diverse SAFs and accommodate various cell demands.
© 2017 Chang et al.
Figures



References
-
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. . 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. 10.1107/S0907444998003254 - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

