RNA-Seq Profiling of Intestinal Expression of Xenobiotic Processing Genes in Germ-Free Mice
- PMID: 28939687
- PMCID: PMC5676297
- DOI: 10.1124/dmd.117.077313
RNA-Seq Profiling of Intestinal Expression of Xenobiotic Processing Genes in Germ-Free Mice
Abstract
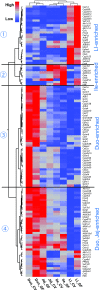
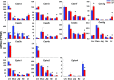
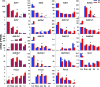
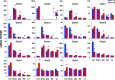
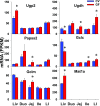
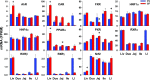
Intestinal bacteria can affect xenobiotic metabolism through both direct bacterial enzyme-catalyzed modification of the xenobiotics and indirect alterations of the expression of host genes. To determine how intestinal bacteria affect the expression of host xenobiotic-processing genes (XPGs), the mRNA profiles of 303 XPGs were characterized by RNA sequencing in four intestinal sections and compared with that in the liver from adult male conventional (CV) and germ-free (GF) mice. Fifty-four XPGs were not expressed in the intestine of either CV or GF mice. The GF condition altered the expression of 116 XPGs in at least one intestinal section but had no effect on 133 XPGs. Many cytochrome P450 family members such as Cyp1a, Cyp2b10, Cyp2c, and most Cyp3a members, as well as carboxylesterase (Ces) 2a were expressed lower in the intestine of GF than CV mice. In contrast, GF mice had higher intestinal expression of some phase I oxidases (alcohol dehydrogenase 1, aldehyde dehydrogenase a1l1 and 4a1, as well as flavin monooxygenase 5) and phase II conjugation enzymes (UDP-glucuronosyltransferase 1a1, and sulfotransferase 1c2, 1d1, and 2b1). Several transporters in the intestine, such as bile acid transporters (apical sodium-dependent bile acid transporter, organic solute transporter α and β), peptide transporter 1, and multidrug and toxin extrusion protein 1, exhibited higher expression in GF mice. In conclusion, lack of intestinal bacteria alters the expression of a large number of XPGs in the host intestine, some of which are section specific. Cyp3a is downregulated in both the liver and intestine of GF mice, which probably contributes to altered xenobiotic metabolism.
Copyright © 2017 by The American Society for Pharmacology and Experimental Therapeutics.
Figures




References
-
- Danielson PB. (2002) The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab 3:561–597. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

